Contributors: Snehal Dilip Karpe, Lingaraja Jena, Malarvizhi A, Santosh Behera, Mahendra Pal Singh, Veronica Medikare, Govardhan Kothapalli, Sidharth Shankar Jha, Jitesh K. Pillai, Uzma Saeed, Puneet Saxena, Chandra Sekhar Pedamallu
Introduction:
In the rapidly evolving field of drug discovery, data has become an invaluable component that drives innovation and efficiency. The advancement in Next Generation Sequencing (NGS) technologies and cost-effective platforms are enabling the generation of enormous amounts of OMICS data in the field of biomedical research. Harnessing these data from diverse sources like genomics, transcriptomics, proteomics, clinical trial data, and real-world evidence (Figure 1), one can uncover new insights, accelerate drug development, and bring effective therapies to patients more quickly. This carefully curated data and associated metadata are called as data assets. However, various data types are stored and present in different data silos and therefore, may not give a holistic view of a drug, disease, or target. On the other hand, an integrated analysis of these data assets does have a wide range of implications for understanding disease, drug mechanisms, and therapeutic targets. While our omics data whitepaper details on the guidance, framework, and benefits of building data assets, the current blog continues to swiftly tap over the scientific value and focusses more on the challenges and alternates to bypass them.
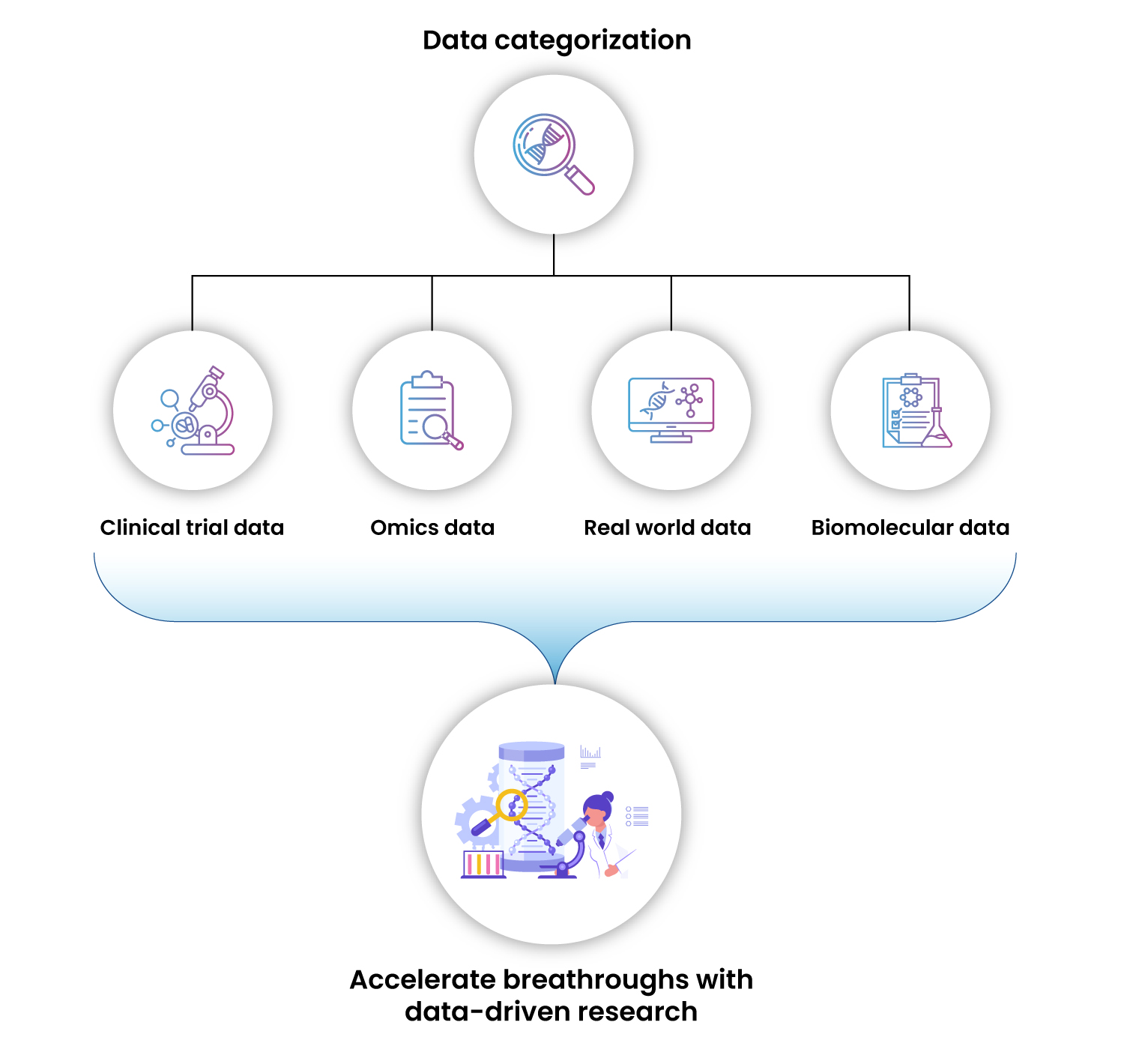
Figure 1 : Types of Data Assets in Biomedical Research
Data assets play a critical role in the advancement of drug discovery research. Integrating OMICS datasets of diverse data types such as genomics, transcriptomics, and proteomics helps researchers to understand disease pathophysiology and decipher drug mechanism of action (MoA). Advancements in data analysis techniques push the application of data asset boundaries to even predict the therapeutic synergies, mechanistic differences between drugs, and drug repositioning opportunities for existing drugs. Through integrated OMICS analysis approach, researchers leverage disease, target, and drug signatures to identify biomarkers that may enhance diagnosis, treatment selection, and drug discovery efforts (Figure 2). All such information, eventually, helps the drug discovery and development process 2.
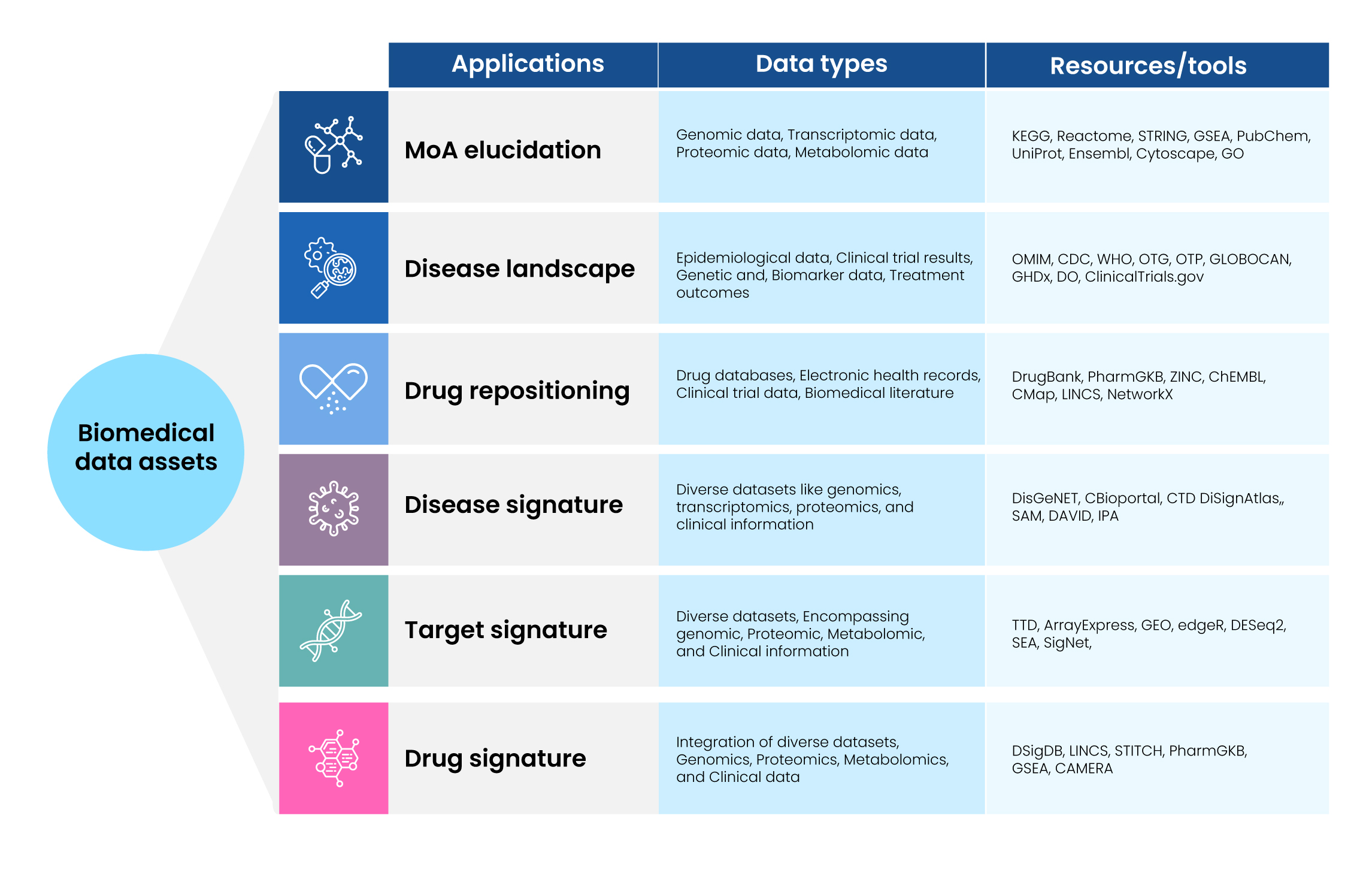
Figure 2 : Applications of Biomedical Data Assets and Associated Resources and Tools
Overall, these biomedical data assets and analysis tools can propel innovation and foster novel treatment discoveries with limited time and budget. These therapies can bring significant improvements to patient care. The key success factors for data assets are data and metadata accessibility and quality. However, in its original form, the data lacks the qualities that make it an “asset” for an organization and many challenges need to be tackled for quality asset creation.
Factors to take into account while building Data asset
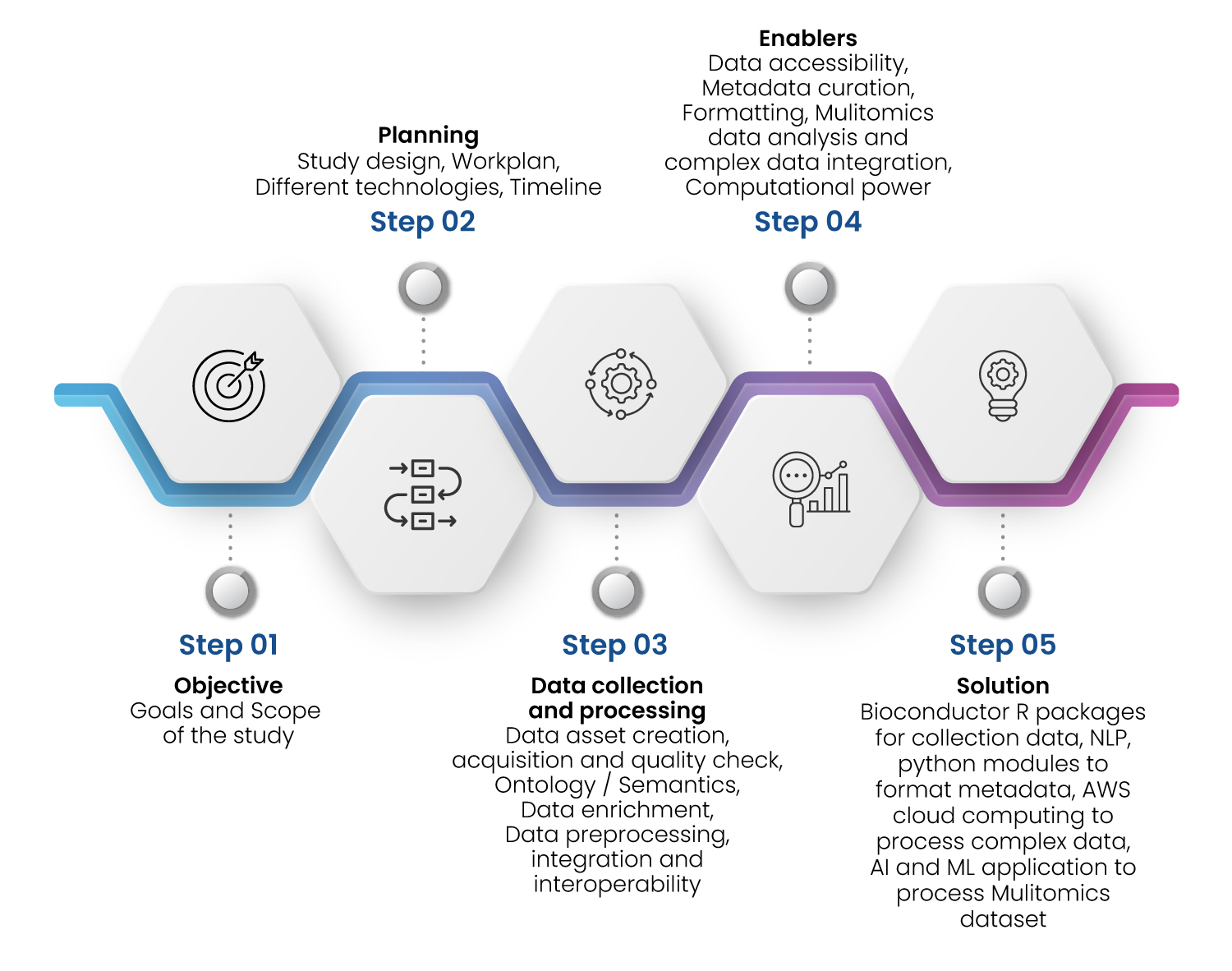
Figure 3 : Excelra’s Approach to build Data Asset
There are multiple challenges one can encounter during the data asset building and integration activity using publicly available datasets that hamper the data quality significantly 3,4. A systematic approach must be followed to handle each of these challenges (Figure 3).
Data quality may be affected by heterogeneity, incompleteness, complexity, redundancy, inconsistency, preprocessing and downstream analysis issues, and issues related to storage for reusability.
Data Heterogeneity: Biomedical data is scattered and heterogeneous in nature. There are multiple factors contributing to this (Figure 4).
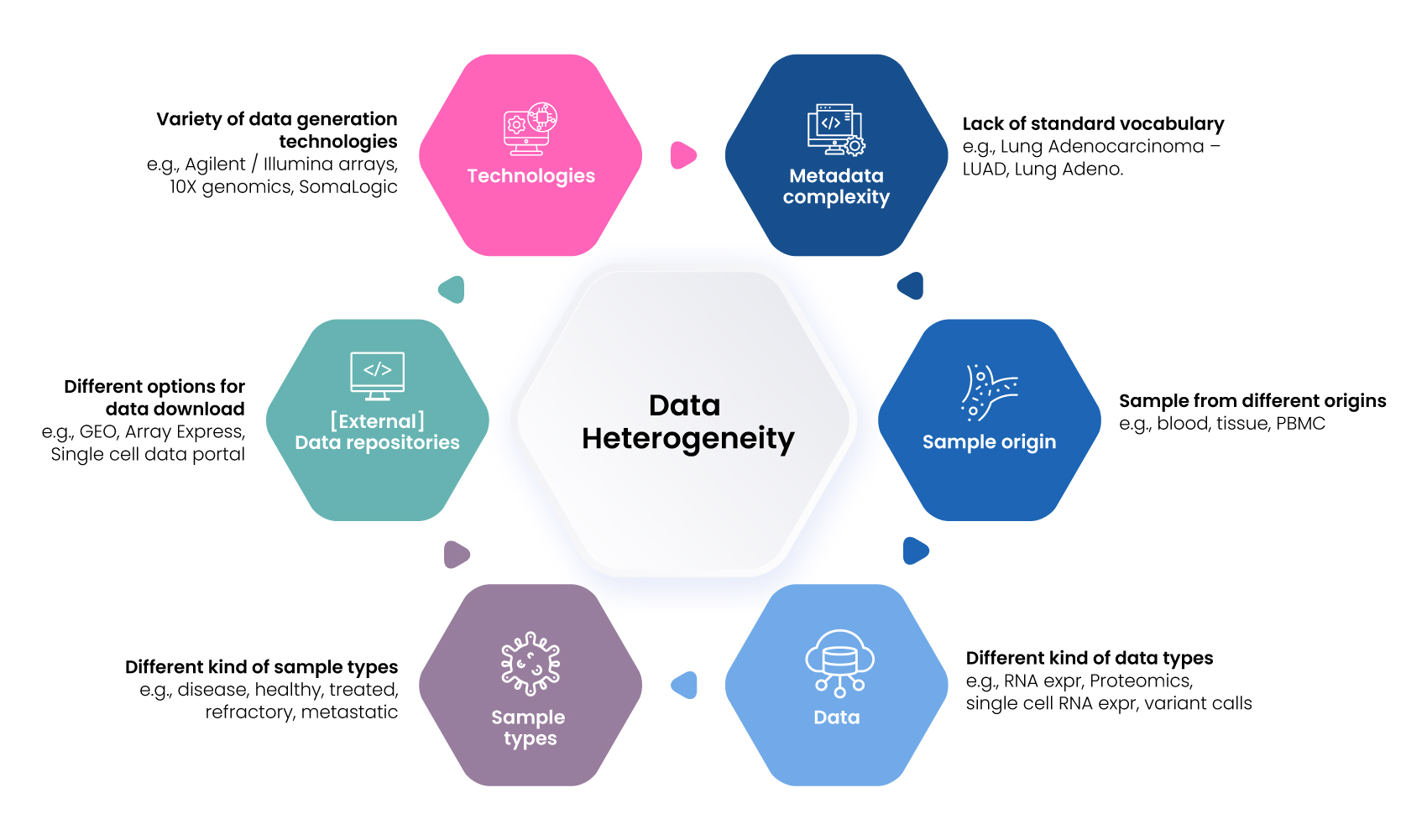
Figure 4 : Factors contributing to Data Heterogeneity
Identification of reliable data sources is the first step of data asset creation. The data sources may be clinical, publications, or open source and may be spread across various databases (Figure 5), each following a specific system of preprocessing, files, formatting, and storage for different kinds of omics data.
Figure 5 : Omics Data Types and Associated Databases
Additionally, some crucial information like the detailed methods of dataset creation, population-level details, individual/sample-level details like disease severity scores, detailed tissue source, and inclusion/exclusion criteria may not be adjacent to the data deposited in the repositories nor can it be easily retrieved using automation. The linked information required to associate individual samples with patients, their omics data, and any other clinical metadata, therefore, needs to be carefully captured by subject matter experts. Data and metadata must be carefully cleaned and validated. Controlled vocabulary, ontology, and semantics principles must be applied to make the information machine readable for further analysis. In addition, extensive documentation of their sources, databases, and SOPs used in the curation process is required. These facilitate scaling up of the process and integration of new features in the data asset whenever required.
Data completeness: Certain data types require extensive information to be useful, e.g. a biomarker has relevance only in the context of the assay performed, quantitative range, and directionality. The kind of analysis and comparisons that can be done on the data is solely dependent on the features captured in the metadata. In such situations, careful planning of the metadata models is required before stepping ahead. Missing data points may be imputed or samples or variables may be removed to ensure that the data could be used for further analysis.
Data harmonization: Correct preprocessing methods are required for each type of omics dataset such that they can all be brought into the same range for further compilation. E.g. The conversion of probe IDs to reference gene IDs (e.g. HGNC symbols) for microarray gene expression data using ID mapping tools / R packages / python modules, usage of the same genome annotation version (e.g. GRCh38) for the bulk RNA-seq data. Missing raw count files is a challenge for the downstream analysis tools like DESeq. Fastq file processing may result in the requirement of increased computational power that can be handled by use of cloud. The presence of data errors, outliers, noise, redundancy, or missing values should be identified through rigorous quality checks and rectified using the latest solutions like fastqc which provides a thorough report on the quality and artifacts, that can be corrected using a plethora of tools.
It is essential to conduct batch effect correction and/or cross-platform co-normalization using advanced statistical or machine learning tools. The selection of apt method/s is a challenge, as it profoundly affects expression values and the outcomes of differential expression analysis, thereby shaping the overall interpretations derived from the data. A tailor-made solution must be identified that is suited for each collection of datasets while ensuring that the final integration is seamless. We can notice and correct issues in the batch-correction using methods like PCA.
Data analysis: Ensuring the retention of the biological signal is intertwined with the selection of an enrichment and integration strategy. There may be hierarchies of information and signals that may be jeopardized if there is a gross generalization. On the other hand, the data asset may become fragmented if we break it into too many categories, resulting in lower sample counts, and making the analysis statistically insignificant. Input from various stakeholders can help to overcome these challenges. Validating the results with the published data can ensure that the processing has worked as intended, however, this may have its challenges as well. The information may be spread across supplementary files or images. There may be inconsistencies, a lack of connection with the datasets deposited in repositories, unmatched contrasts, and incomplete or outdated methodologies, parameters, and statistical tests. Hence subject matter expertise and manual efforts may be required to acquire the correct information. Appropriate measures need to be taken to avoid issues while classifying rare events due to class imbalance and overfitting due to the curse of dimensionality while applying ML tools to these data assets.
Cross-omics Integration: A classical analysis framework includes separate analysis of each kind of omics data followed by the late integration of the obtained results. Early integration is now possible with the availability of meta-analysis methods and other algorithms 5. However, the challenges mentioned before may compound with additional difficulties in understanding the results of the multi-omics models in terms of mechanistic insights, novel regulatory mechanisms, and multi-layer data visualization issues.
Data Ingestion: There are tools available to enhance data asset storage, analysis, and visualization like TileDB and Rosalind that simplify usage by the larger community, however, there may be several questions encountered at each analysis step E.g. what to do in case of small sample sizes, how to define outliers and should we remove them, how to correlate and integrate results from two different technologies (like microarray and RNAseq), etc. A subject-matter expertise and omics data analysis knowledge would be crucial to find solutions to such questions.
Finally, a few scientific questions may be addressed only using AI/ML approaches, deep learning, NLP, etc. This is typically the larger goal of creating huge data assets. Hence, the data asset should be prepared in such a way that using it as an input, or extracting features from it and running further computations is a smooth process.
Use case:
Biopharma companies are often interested in indication prioritization for their novel drugs. This helps them to expand their portfolio and increase the life cycle of their drug. Developers are also interested in patient stratification into responders, partial responders, and non-responders in the disease of interest. Typically, this requires costly, time-consuming experiments or clinical trials. Instead, the identification and creation of data assets from publicly available sources and their analysis may identify potential patient stratification biomarkers. This approach of using data assets and analytics can drastically reduce the time and resource requirement for pharma companies (Figure 6).
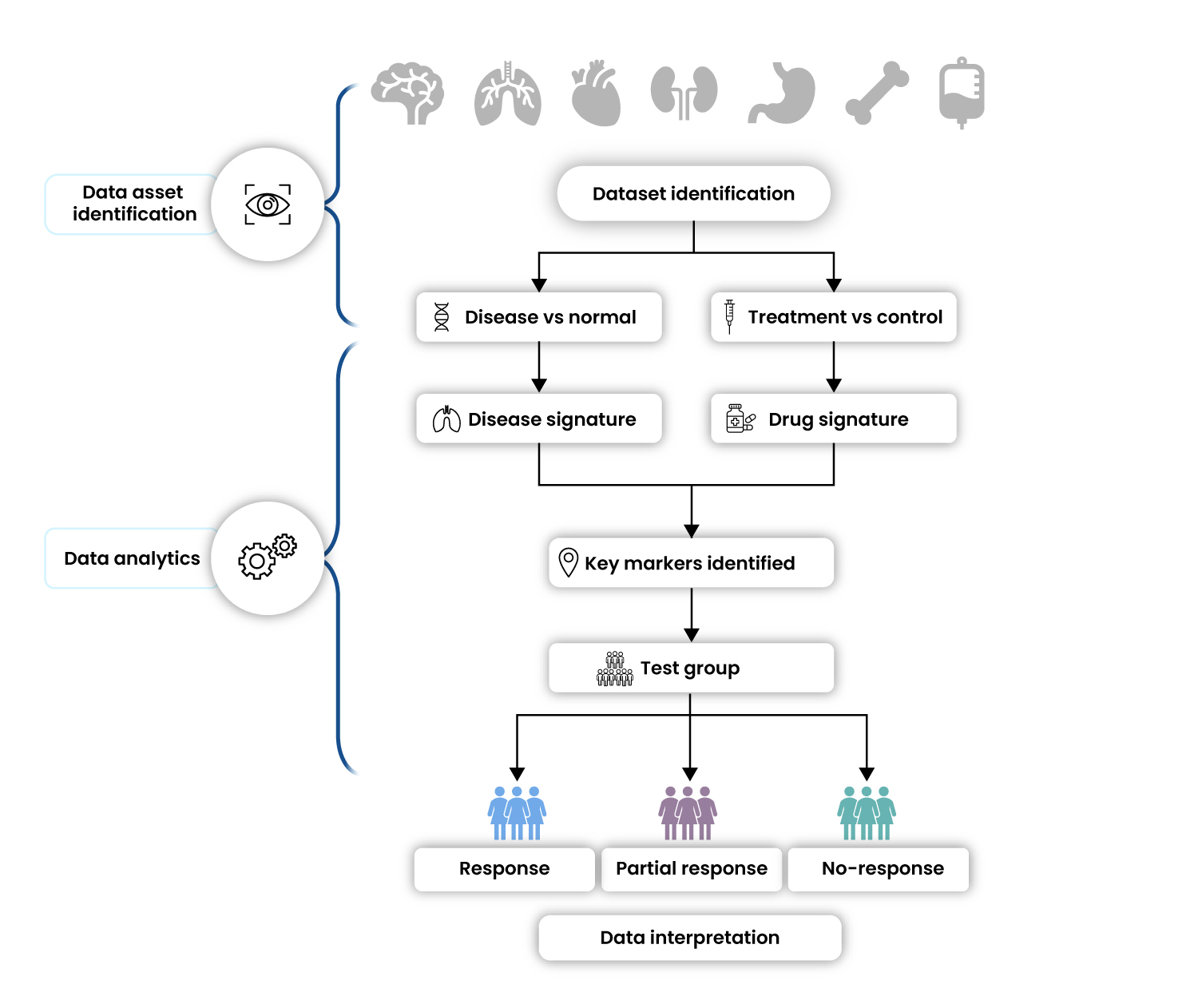
Figure 6 : Data Asset Creation for Patient Stratification
As the percentages of responders to the novel asset will not be available in the public data for the indications under study, it would be necessary to identify a proxy for that. Transcriptomics data captures the global gene expression status of the samples under study and can harbor signals that indicate sensitivity to a specific treatment. An analysis framework can be defined to study transcriptomes of diseased and healthy individuals from all available public datasets, identify the molecular anomalies seen in the diseased samples, compare them with the mechanism of action or the known signatures of the drug effect, and identify diseased individuals who are likely to respond better to the given drug.
According to the framework in our Whitepaper, a focused objective is defined. The framework of analysis depends on the global gene expression, so the scope of the study could be limited to microarray and bulk RNA-seq datasets. A team of subject matter experts identified relevant datasets and gathered data and metadata from the public repositories. As per the next steps of the framework and best practices mentioned in the Whitepaper, the process required data enrichment, interoperability, and integration. Analysts tackled intermittent challenges case-by-case basis as discussed before.
Since the datasets were collected from a diverse set of studies, they had to be harmonized based on compatible groups. Finding the best method for the purpose without reducing the biological signal and relevancy of the dataset is one of the biggest challenges. One of Excelra’s USPs is our pool of multi-omics analysis experts that are well-versed with implications arising from different normalization / batch-correction algorithms and validation processes (Figure 7).
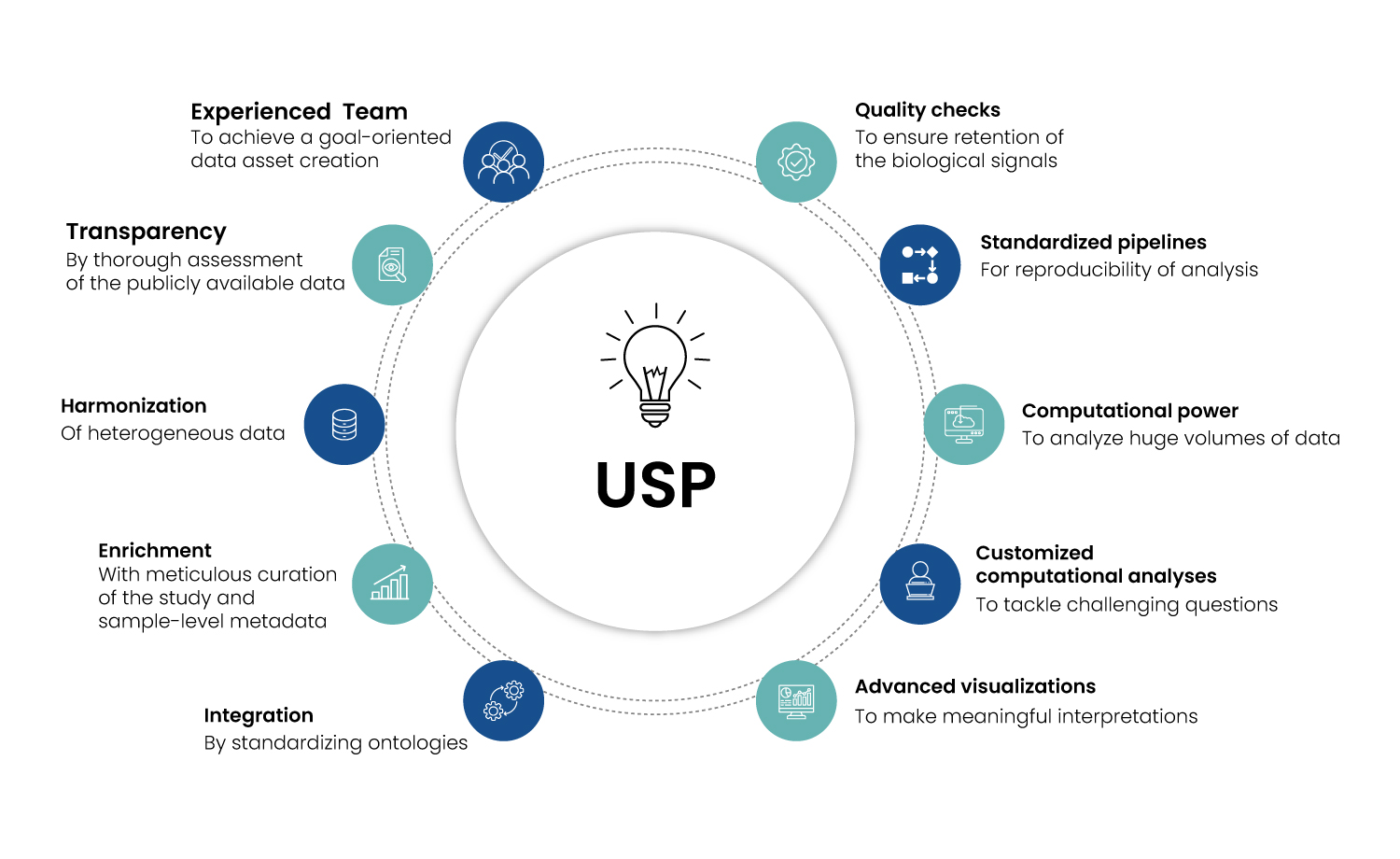
Figure 6 : Excelra’s Unique Selling Points for Biomedical Data Asset Creation
As per the plan, the raw and harmonized data and analysis results were stored in specific formats suitable for further ingestion.
Once harmonized data was ready, a comparison of the signatures that capture the drug’s effects across healthy and diseased individuals was performed for one of the shortlisted diseases. The individuals, with drug effect signatures displaying drastic deviation from the healthy samples, could be hypothesized to respond better to the treatment as the drug would likely push the trends towards the healthy patterns. This helped in response stratification and provided a drug-response score to each individual. This was repeated for other indications and hence assisted in highlighting the indications with a higher number of responders that may be prioritized for further clinical trials with the drug asset. This way, careful data asset creation from public datasets could be leveraged to generate novel insights for the drug asset of interest.
A way forward / future prospects:
The modern drug discovery process is increasingly becoming data-driven. Organizations are investing billions in big data analytics, AI-based innovation, and overall digital transformation. Meanwhile, High costs and data complexity challenge drug discovery, making the efficient use of expensive experimental and computational data crucial. Although the process is fraught with multiple challenges, there are many standardized solutions that are becoming available. Biomedical data assets are revolutionizing drug discovery and translational research by effectively managing, storing, and analyzing vast amounts of complex data. Data asset creation has thus become crucial to maintaining the competitive edge in today’s Pharma and Biotech world.
Excelra offers customized solutions to create valuable biomedical data assets from publicly available data. To know more about our offerings please reach out to us


