Introduction
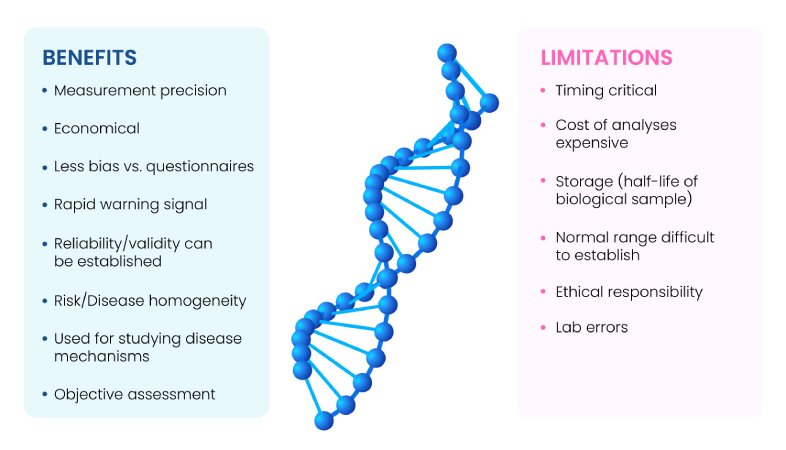
Biomarkers are indicators of homeostasis, disease states, or pharmacological interventions. An ideal biomarker must be binary (present or absent), quantifiable in tissues, cells, body fluids, or in simple physiological parameters such as blood sugar or blood pressure with minimal variability, should have substantial signal to noise ratio (sensitive and specific), and vary promptly and reliably in response to changes due to an underlying condition or treatment. The main benefits and limitations of biomarkers are summarized in Fig. 1(1).
Fig 1: Benefits and limitations of Biomarkers
Also referred to as biological markers, molecular markers, surrogate endpoints, or signature molecules, biomarkers are of immense significance to biopharma, diagnostics, personalized medicine, and overall healthcare industry. The global biomarkers market valued at USD 66.97 billion in 2022 is expected to grow at a CAGR of 13.3% from 2023 to 2030(2). Variables such as enhanced impact of companion diagnostics, rise in incidence and prevalence of cancers, cardiovascular, neurological, and immunological diseases, higher private and public investments and funding, noteworthy inventions from current R&D, and novel digital health technology solutions are expected to disrupt and catapult the growth of the biomarker industry(2).
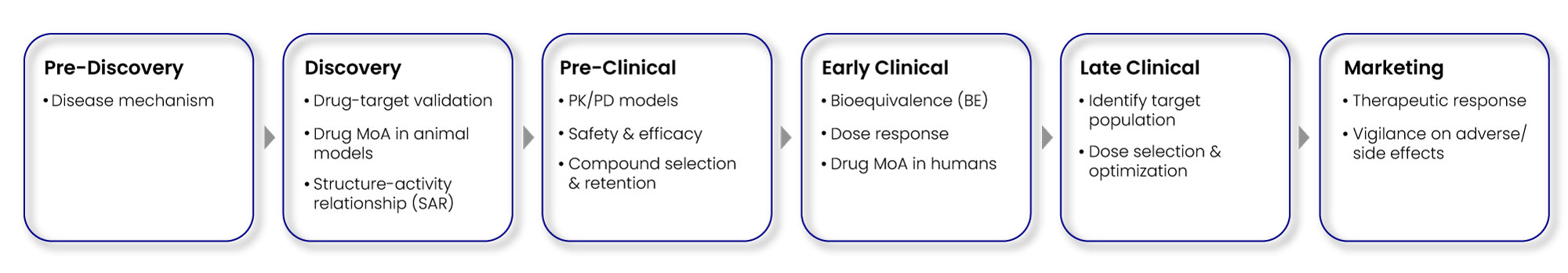
Biomarkers have a wide spectrum of functionalities, ranging from early drug discovery, to pre-clinical, and finally late-stage clinical development as depicted in Fig. 2.
Fig 2: Diverse biomarkers in drug discovery & development process
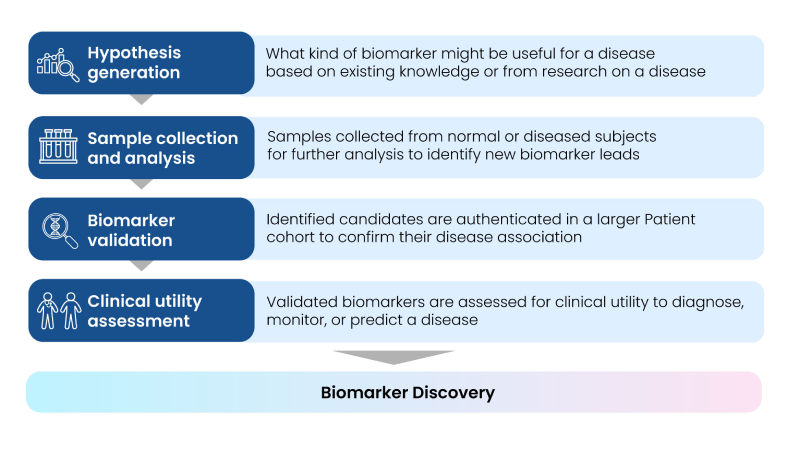
The process of biomarker discovery involves identifying, measuring, and validating proteins, peptides, hormones, or DNA/RNA in biological samples, and finally selecting potential candidates for various R&D applications. This process is imperative to identify a drug’s mechanism of action, investigate toxicity and efficacy signals during early development, measure drug pharmacokinetics/pharmacodynamics (PK/PD), and identify patients who are likely to respond to specific therapies. In the early stages of drug development, biomarkers may also help identify disease-specific molecular pathways(3).
Using specific and reliable biomarkers can ensure efficient and streamlined drug discovery, development, regulatory and approval processes, facilitate design and production of effective drugs, patient stratification for clinical trials, and benchmarking of disease endpoints and health outcomes(4).
The entire workflow of profiling disease-specific biomarker leads is summarized in Fig. 3.
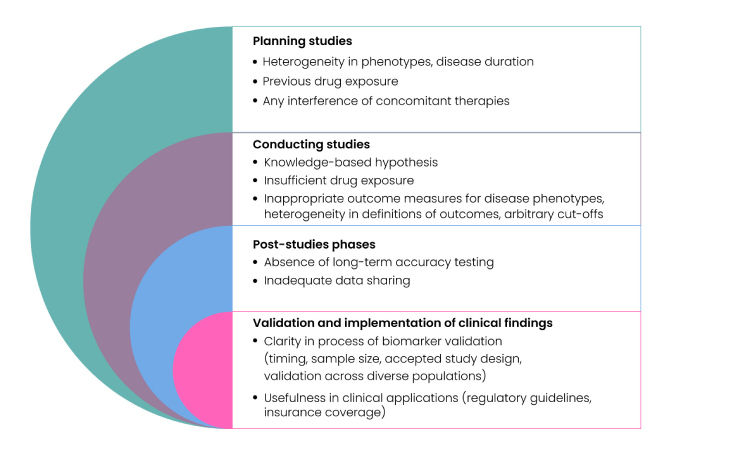
The process of biomarker discovery, validation, and qualification has several challenges. Inadequate patient selection and recruitment, small sample size, poor sample handling, limited metabolomic and proteomic data, poor study designs, and inadequately defined cut-offs for measuring biomarkers are a few rate limiters as shown in Fig. 4.
Fig 4: Challenges in Biomarker Discovery
Multiple variables such as age, race, gender, and lifestyle can make it difficult to discern actual effects of a disease as opposed to direct effects of such demographic or clinical factors. Moreover, the process can be slow, expensive, time- and resource-intense, often requiring large studies across multiple populations and ethnicities, with long follow-up periods, thereby making it difficult to obtain enough funding for biomarker discovery research. Also, limited precision of biomarker assays, lack of standardized procedures, poor characterization of diagnostic accuracy can make the precise clinical utility of biomarkers confounding; while some are useful for diagnosis, they may simultaneously not be suitable for monitoring disease progression or for predicting therapy response(5). Recent technological advances in multi-omics, cytometry, and imaging approaches, combined with novel bioinformatics and biostatistics methodologies can circumvent some of the current challenges in traditional biomarker discovery and accelerate the discovery and development of reproducible biomarkers for complex diseases(3).
Background
Biomarker landscape assessment
The previous section referred to the long, arduous trajectory of biomarker discovery. Assessing the landscape of biomarkers is imperative to evaluate their prospective functional, strategic, and commercial impact and categorization in drug discovery and development, disease detection and monitoring, while ensuring sufficient coverage of drug-target-disease associations. Diagnostic biomarkers can detect a disease before clinical symptoms appear while prognostic biomarkers like HER2 overexpression or amplification can predict future disease outcomes in breast cancer patients along with overall survival rate. PSA levels or Gleason scores serve as predictive biomarkers to indicate potential disease risk and measure therapeutic response in multiple patient populations. A few examples of functionally distinct biomarker types are provided in Fig. 5.
Fig 5: Functionally distinct biomarker types
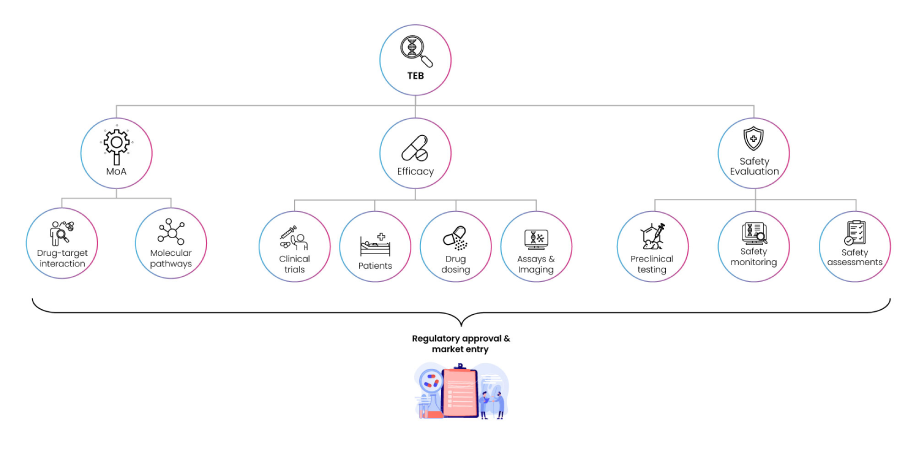
To bridge bench-to-bedside efforts, translational biomarkers are used for identifying the impact of a clinical compound on body systems (organs, tissues) much before a specific clinical effect is observed. During enrolment in a clinical trial, safety biomarkers on the other hand can predict the probability that a patient might experience adverse events or toxicity due to specific drug treatments. Overall, clinical biomarkers encompass the entire spectrum of biomarker functionality – from disease diagnosis, monitoring progression, or predicting treatment course and responses in the right patient groups. Amongst clinical biomarkers, Target engagement biomarkers (TEBs) are used for confirming if drug-target binding creates a biological response, and if so, the optimal dose at which this can occur with minimal on- and off-target side effects(6).
Fig 6: Functions of Target Enhancement Biomarkers (TEBs)
TEBs monitor In vivo efficacy, safety, and drug resistance, and PK/PD properties of promising compounds in pre-clinical, genetically modified or xenograft animal models of specific diseases(7; 8; 9) as shown in Fig. 6. Linking TEBs and mechanistic PK/PD models can reveal underlying mechanisms of drug action, establish relationship between dose and biomarker response, assess impact of dose escalation studies on time course of response, measurement of response duration and lag between plasma drug concentration and response, measurement of effect of repeated doses (tolerance, sensitisation, reflex mechanisms), range of doses, dose separation, and dosing interval to form basis of clinical trial simulations, paving the way to make informed go/no-go decisions for designing future clinical studies.
Regardless of how biomarkers are functionally defined or classified, their development and application should be determined by the fit-for-purpose principle in multiple physiological and pathological states(3). With the advent of digital transformation, high-performance cloud computing, cloud data storage, management and automation, a new era has begun where disruptive and innovative technologies such as AI/ML are re-defining the value proposition of biopharma, healthcare products and services, and life science research.
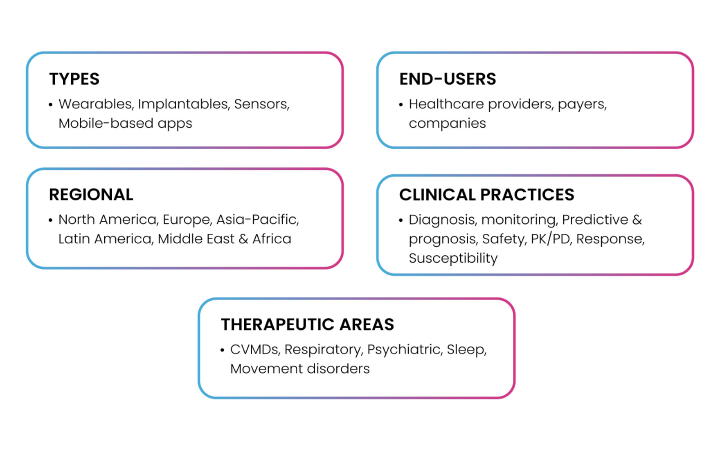
For instance, digital biomarkers can potentially act as a bridge between traditional medicine and digital health and therapeutics. In the aftermath of the COVID-19 pandemic, there has been a steep rise in digital health practices, mostly due to a surge in healthcare costs, deteriorating health outcomes, worsening incidence of respiratory, infectious, and cardiovascular conditions, a sharp rise in the prevalence and accessibility to mobile- and electronic health (mHealth and eHealth), IoT, and wearable technologies(10). The FDA defines a digital biomarker as a characteristic or set of characteristics collected from digital health technologies (DHTs), measured as an indicator of normal biological or pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions. These biomarkers hold immense potential in the process of detection and selection of ideal biomarker candidates and can provide massive volumes of data to reveal population health trends, illness patterns, risk factors, and treatment outcomes. Biogen, Amgen, Apple, Altoida, AliveCor, GlaxoSmithKline, Kintsugi are some of the well- known players who are revolutionizing the digital biomarker landscape. Fig. 7 depicts the current market segmentation for digital biomarkers based on types, end-users, geographies, utility in clinical practices, and specific therapeutic areas of implementation.
Fig 7: Digital Biomarkers Market Segmentation
Behavioural and physiological data such as heart rate, physical activity, sleep patterns, body temperature, blood pressure, and step counts collected on digital wearable devices like fitness trackers or smartphones that incorporate eHealth/mHealth apps and software, computing platforms, internet connectivity, and sensors for healthcare and related users can be considered as effective digital biomarkers. A library of more than 430 digital endpoints, sensors/devices used to measure them, and related clinical trial information is catalogued by the Digital Medicine Society(11) to facilitate discovery of digital biomarkers.
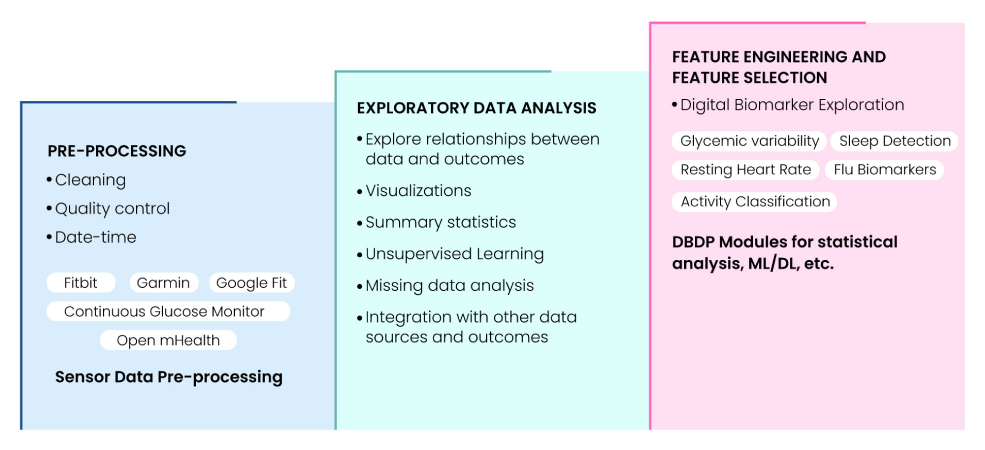
The Digital Biomarker Discovery Pipeline (DBDP) is the first comprehensive, open-source software platform using FAIR principles for end-to-end digital biomarker development(12). DBDP modules assist in calculation and utilization of biomarkers like resting heart rate (RHR), glycemic variability, insulin sensitivity status, exercise response, inflammation, heart rate variability, activity, sleep, circadian patterns, and flu biomarkers to predict health outcomes. It also provides resources for Digital Biomarker Discovery Education, open DBDP in collaboration with Open mHealth, and Digital Health Data Repository (DHDR). Fig. 8 summarizes the components of the digital biomarker discovery pipeline that can be handled by DBDP.
Fig 8: Digital Biomarker Discovery Pipeline (DBDP) landscape
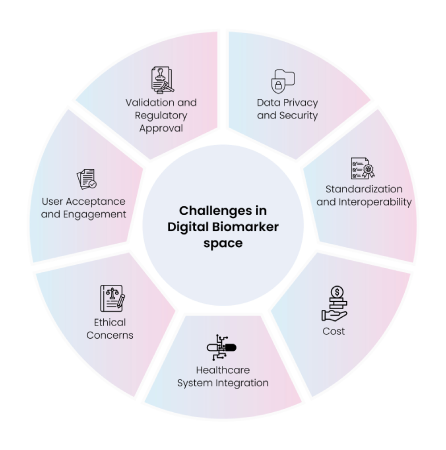
Although interest in digital biomarkers is evolving rapidly, the associated real-world data is not easily available, hindering efficient use of tools and technologies required for digital health data analysis. A lack of regulatory oversight, limited funding opportunities, general mistrust of sharing personal data, and shortage of open-source data and codes are some of the challenges linked to digital biomarker development. Safeguarding sensitive health information and ensuring HIPAA and GDPR compliance are also critical. The process of transforming digital data into digital biomarkers is computationally expensive, along-with a dearth of standard validation methods. Fig. 9 summarizes the key challenges associated with developing digital biomarkers.
While one-size-fits-all approach to identify and select traditional or digital biomarkers may not always be feasible, it has become imperative to identify and use new approaches for biomarker profiling based on data specificity, development cost, process complexity, scope, efficiency and radical power of the technologies or molecular tools(13; 14).
Trends/Technologies in biomarker discovery
While choosing the correct target-disease-drug data is a mandate for biomarker discovery, an abundance of unstructured, siloed, and non-FAIRified (Findable, Accessible, Interoperable, Reusable) data is a major impediment to the data cleaning, fusion, and integration efforts of both the traditional and recent methods/tools used in identifying and discovering ideal biomarker candidates. Fig. 10 highlights the different tools and methods used and their benefits and drawbacks with respect to parameters such as specificity, efficacy, cost, time, and efforts.
Fig 10: Comparison of traditional and vs. recent methods/tools in biomarker discovery
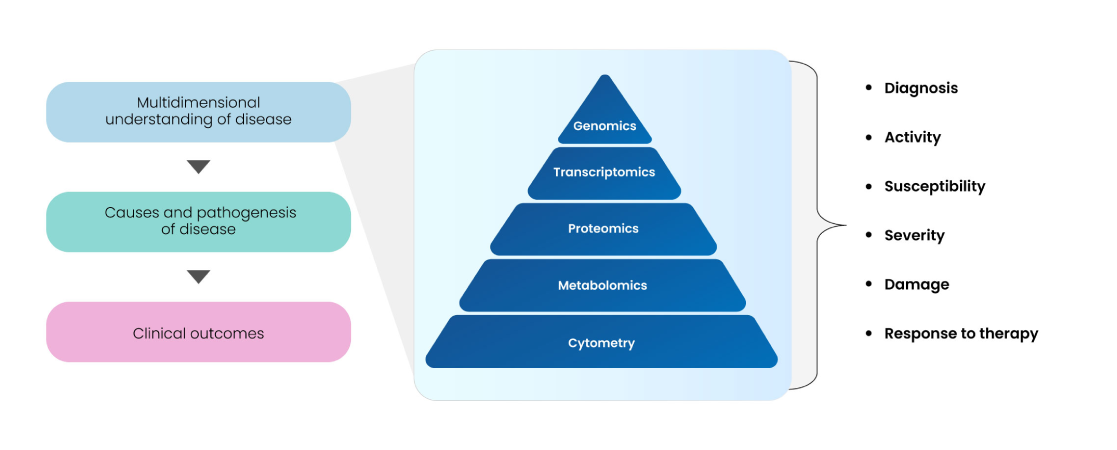
As depicted above, multi-omics, cytometry, and imaging technologies are amongst the most promising technologies today to decipher various facets of target-disease-drug associations, along-with multivariate data types, databases, bioinformatics, and biostatic analytical tools. How they impact biomarker discovery and validation, aid in understanding disease biology, and engender robust clinical translatability have been depicted in Fig. 11.
Fig 11: Multi-omics approaches for biomarker discovery and validation
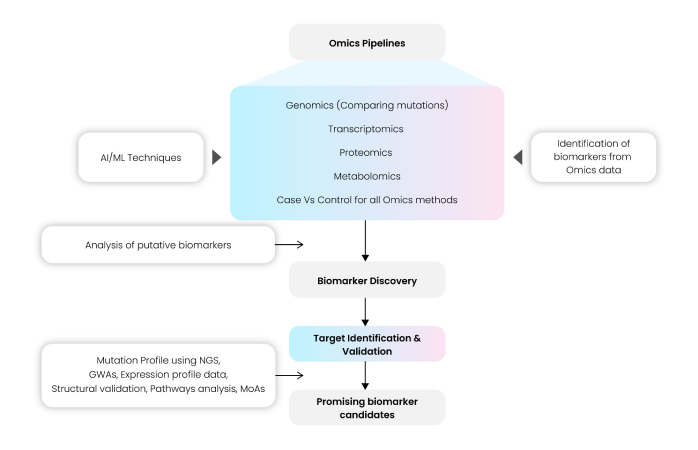
Different genomic biomarkers (gene mutations or polymorphisms), transcriptomic biomarkers (gene or miRNA expression profiling), allele/haplotype mapping, epigenomics, SNP analysis, pharmacogenomics, proteomics, metabolomics, glycomics, and other small molecules can be used as end-point markers of better health outcomes and clinical translation. Tailoring multi-omics pipelines based on specific data types and integration of AI/ML algorithms can facilitate the discovery of novel targets and subsequently categorize them as prospective biomarkers. The schema in Fig. 12 provides an overview of the typical workflow of an Omics platform used for biomarker discovery.
The pipeline includes several important steps to perform quality control and validation of input data files (gene expression or proteomic data) using parameters such as Phred score, Library size and presence of adapter sequences. Normalized data used for further processing, downstream analysis and statistical methods for target identification and validation can be selected based on most used standard tools and methods in scientific literature. Further-on, proteomic data are used to validate the putative candidates, along-with statistical methods and correlation studies. Mutation profiling, GWAS analysis, pathway analysis, determining potential mechanisms of action help in finalizing and selecting the top biomarkers. However, these biomarker measurements come with their limitations in statistical methods, data normality and distribution, outliers and undetected biomarker limits, or gene duplications and selection, computational complexity, and high-dimensional datasets(17).
Fig 12: Omics pipelines used in Biomarker Discovery
The development of machine learning (ML)-based methods can circumvent some of the challenges linked to scale, diverse data distributions, and non-linearity. ML algorithms can identify patterns in large datasets to predict outcomes or classify groups based on input data(17).
Impact of Machine Learning (ML) on biomarker discovery
The deluge and availability of large annotated multi-omics data and advances in computing power have prompted the implementation of artificial intelligence and machine learning (AI/ML) algorithms in early drug discovery and development. The rapid advances in AI/ML and deep learning (DL) are facilitating prediction of complex drug response and resistance based on different types of genomic, transcriptomic, proteomic and metabolomic input data(15), as depicted in Fig. 13.
Fig 13: Input data types used in ML-based biomarker discovery
The prediction outcomes can be capitalized on to identify and validate novel biomarker candidates. Applying supervised or unsupervised ML/DL ensemble models on biomedical data can eventually accelerate development of personalized therapies, drug repurposing, drug discovery efforts, biomarker development, and aid in improving decision-making, diagnosis, and treatment at the point-of-procedure(16).
Fig 14: General ML workflow for biomarker identification
Data pre-processing is a crucial step in this ML-based biomarker discovery. The pre-processed data is then transformed for use in building and training specific ML models(17). Fig. 14 outlines the key steps executed through the ML approach for biomarker discovery. The most common ML algorithms that have successfully been used for biomarker discovery are Support Vector Machine (SVM), Random Forest (RF), K-Nearest Neighbour (KNN), Logistic Regression (LR), Cox regression, LASSO regression, ANOVA and Deep Neural Networks(18).
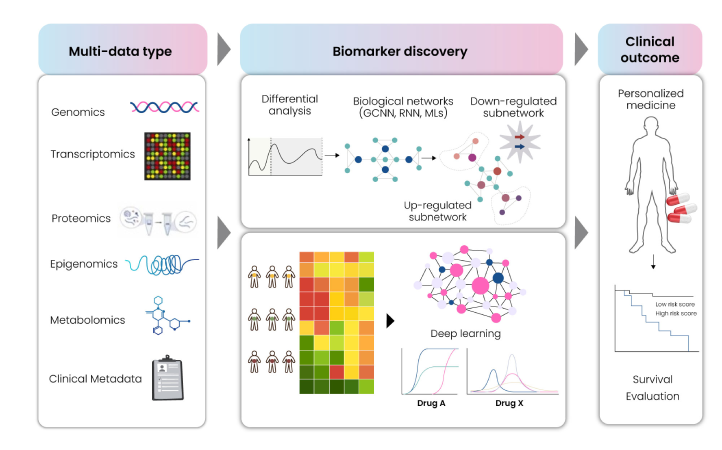
The initial step of model training for drug response prediction consists of generating patient-specific features aiming towards identifying biomarker candidates based on the predicted response of a patient to the real world/ simulated drug treatment. The features generated by the scoring algorithm in the ML/DL model may need modification and re-evaluation of the class assignment of the patient may be required for efficient training of the model. After re-evaluating the predictions made by the ML/DL model, a ranking to promising biomarkers by the proportion of disease samples can be classified as normal or as a proxy of the effectiveness of the drug. The trained ML/DL model must learn to accurately differentiate between normal and disease samples by modifying the features or changing the biomarkers. For validation, identified biomarker-drug pairs predicted as true positives can be verified by mapping to reference datasets like Genomics of Drug Sensitivity in Cancer (GDSC), FDA-approved drugs, ClinicalTrials.gov etc. Fig. 15 depicts an overall ensemble workflow for biomarker discovery based on deep learning (DL).
Fig 15: DL-based ensemble workflow for Biomarker Discovery
Conclusion/Outlook
In terms of productivity, the biopharma sector has its high and low phases, with failure rates in clinical trials over 90% despite pre-clinical testing, and costs incurred to develop new drugs exceeding $2.6 billion(16). Despite numerous pitfalls, the significance of biomarker discovery in disease diagnostics and therapeutic monitoring is progressing incrementally. The current whitepaper has endeavoured to bring out the diversity of this topic, traveling across the landscape of various types of molecular biomarkers, trends in their R&D applicability, and delved into the unprecedented usefulness of novel disruptive tools and technologies, heralding data-driven approaches to biomarker identification, selection, verification, and validation. The integration of AI/ML approaches, digital transformation, and adherence to ethical and regulatory standards open the floodgates to harness new strategies for advancing precision and personalized medicine and enhancing patient health outcomes using conventional as well as digital biomarkers. With newer advances in AI subtypes such as generative AI(19) and large language models (LLMs) such as ChatGPT(21), AI is bound to become a force to reckon with, especially for mining large-scale text datasets for identifying novel disease-associated biomarkers, enabling more accurate diagnoses and targeted therapies. While one must tread the path of LLMs cautiously owing to chances of increased inaccuracies and misleading information, their benefits as aids to biomarker discovery and development remain undoubtedly high and at the forefront of innovation in precision medicine and drug discovery, development, and repurposing (22). Concrete strides using AI/ML are thus underway to secure validated and viable solutions in healthcare and research development.
References
1. Dahham Alsoud, Séverine Vermeire, Bram Verstockt. Biomarker discovery for personalized therapy selection in inflammatory bowel diseases: Challenges and promises, Current Research in Pharmacology and Drug Discovery, Volume 3, 2022, 100089, ISSN 2590-2571, https://doi.org/10.1016/j.crphar.2022.100089
2. Biomarkers Market Size, Share & Trends Analysis Report. Report ID: 978-1-68038-979-1: https://www.grandviewresearch.com/industry-analysis/biomarkers-industry
3. Attila A. Seyhan. Biomarkers in drug discovery and development. 2010: https://www.europeanpharmaceuticalreview.com/article/4357/biomarkers-drug-discovery-development/
4. Califf RM. Biomarker definitions and their applications. Exp Biol Med (Maywood). 2018 Feb;243(3):213-221. doi: 10.1177/1535370217750088. PMID: 29405771
5. Landry, Vivianne & Coburn, Patrick & Kost, Karen & Liu, Xinyu & Li-Jessen, Nicole. (2022). Diagnostic Accuracy of Liquid Biomarkers in Airway Diseases: Toward Point-of-Care Applications. Frontiers in Medicine. 9. 10.3389/fmed.2022.855250
6. Paweletz CP, Andersen JN, Pollock R, Nagashima K, Hayashi ML, Yu SU, et al. (2011) Identification of Direct Target Engagement Biomarkers for Kinase-Targeted Therapeutics. PLoS ONE 6(10): e26459. https://doi.org/10.1371/journal.pone.0026459
7. Tang et al., Use of in vivo animal models to assess pharmacokinetic drug-drug interactions, Pharm Res., 2010, 27(9), 1772-1787
8. Storey et al., A Structured Approach to Optimizing Animal Model Selection for Human Translation: The Animal Model Quality Assessment., ILAR J, 2021, 62(1-2), 66-76
9. Long et al., Drug discovery oncology in a mouse: concepts, models and limitations, Future Sci OA., 2021, 7(8)
10. Vasudevan, S., Saha, A., Tarver, M.E. et al. Digital biomarkers: Convergence of digital health technologies and biomarkers. npj Digit. Med. 5, 36 (2022). https://doi.org/10.1038/s41746-022-00583-z
11. Digital Medicine Society: https://dimesociety.org/
12. Bent B, Wang K, Grzesiak E, Jiang C, Qi Y, Jiang Y, Cho P, Zingler K, Ogbeide FI, Zhao A, Runge R, Sim I, Dunn J. The digital biomarker discovery pipeline: An open-source software platform for the development of digital biomarkers using mHealth and wearables data. J Clin Transl Sci. 2020 Jul 14;5(1): e19. doi: 10.1017/cts.2020.511. PMID: 33948242
13. Cui W, Duan Z, Li Z, Feng J. Assessment of Alzheimer’s disease-related biomarkers in patients with obstructive sleep apnea: A systematic review and meta-analysis. Front Aging Neurosci. 2022 Oct 13;14:902408. doi: 10.3389/fnagi.2022.902408. PMID: 36313031
14. Scaros O, Fisler R. Biomarker technology roundup: from discovery to clinical applications, a broad set of tools is required to translate from the lab to the clinic. Biotechniques. 2005 Apr; Suppl:30-2. doi: 10.2144/05384su01. PMID: 16528921
15. Zhang X, Jonassen I, Goksøyr A. Machine Learning Approaches for Biomarker Discovery Using Gene Expression Data. In: Helder I. N, editor. Bioinformatics [Internet]. Brisbane (AU): Exon Publications; 2021 Mar 20. Chapter 4. PMID: 33877765
16. Zhavoronkov A. Artificial Intelligence for Drug Discovery, Biomarker Development, and Generation of Novel Chemistry. Mol. Pharmaceutics 2018, 15, 10, 4311–4313. Publication Date:October 1, 2018. https://doi.org/10.1021/acs.molpharmaceut.8b00930
17. Ng, S., Masarone, S., Watson, D. et al. The benefits and pitfalls of machine learning for biomarker discovery. Cell Tissue Res 394, 17–31 (2023). https://doi.org/10.1007/s00441-023-03816-z
18. Dhillon, Arwinder & Singh, Ashima. (2018). Biology and Today’s World Machine Learning in Healthcare Data Analysis: A Survey. 10.15412/J.JBTW.01070206
19. Generative AI in healthcare & life sciences – revolutionizing healthcare through AI-powered innovation: https://www.linkedin.com/pulse/generative-ai-healthcare-life-sciences-revolutionizing-through-lhj4c
20. Elsborg J, Salvatore M. Using LLMs and Explainable ML to Analyze Biomarkers at Single-Cell Level for Improved Understanding of Diseases. Biomolecules. 2023 Oct 12;13(10):1516. doi: 10.3390/biom13101516. PMID: 37892198
21. Dipesh Uprety MD, Dongxiao Zhu PhD, Howard (Jack) West MD. ChatGPT—A promising generative AI tool and its implications for cancer care. Cancer. 14 May 2023. https://doi.org/10.1002/cncr.34827
22. Cascella M, Montomoli J, Bellini V, Bignami E. Evaluating the Feasibility of ChatGPT in Healthcare: An Analysis of Multiple Clinical and Research Scenarios. J Med Syst. 2023 Mar 4;47(1):33. doi: 10.1007/s10916-023-01925-4. PMID: 36869927
How can we help you?
We speak life science data and help you unlock its potential.