Introduction
Biomarkers are defined as a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention [1] There are more than 40,000 biomarkers being explored in pre-clinical and clinical studies for a broad range of diseases and therapeutic areas. [2], [3] Of that number, however, less than 400 biomarkers qualify through either FDA/EMA approval, presence in drug labels, guidelines or as companion diagnostics. That means 99% of the biomarkers that are actively studied will not impact a treatment’s regulatory approval or a patient’s treatment decision. [4]–[9] The gap between exploratory biomarkers and qualified biomarkers is striking. It is imperative that R&D teams reduce that gap by making informed decisions about which biomarkers to include in their research and pursue for approvals.
A possible solution is biomarker validation. By observing multiple independent references, it’s possible to measure the validity of biomarkers for a defined context of use [10], [11] This white paper will discuss the ways in which multiple contributing criteria can be used to determine a validity index. Referencing the specific example of colorectal cancer, it will show how such an index can be used to grade and prioritize biomarker drug development, aiding in ultimately improving the accuracy of drug trials, clinical treatment and achieving regulatory approval.
Impact of biomarkers and challenges to qualification
The concept of biomarkers can be traced back to ancient times, although the understanding and application of biomarkers as we know them today have developed more recently.[12], [13] Over centuries, an individual’s condition was determined based on their physiological markers, such as pulse rate or body temperature. The developments in medical technology have enabled more quantitative assessments of a multitude of physiological markers. The term “biomarker” itself emerged in the 1980s when researchers began to recognize the broader potential of using specific molecules or biological characteristics as indicators of disease, physiological state, or response to treatment. This coincided with advancements in molecular biology, genetics, and technology, enabling the identification and measurement of a wide range of biomarkers, including genetic markers, proteins, metabolites, and imaging markers. The application of biomarker measurements has ever since expanded beyond disease diagnosis to prognosis, risk, and drug response.
Biomarker measurements remain one of the key activities in medical practice. General practitioners, consultants, and surgeons rely on accurate biomarker measurements in disease diagnosis, treatment and dosing decisions, monitoring efficacy of a prescribed treatment as well as risk assessment.
Pharmaceutical and biotech companies utilize biomarkers at every stage of the drug discovery and development process. The markers are used to help identify drug targets, study mechanism of action (MoA), ADME and safety readouts, measure drug response, and support patient selection and stratification. [12], [13] References to biomarker activity in clinical trials have increased dramatically in recent years, and have had a major impact on drug development programs and regulatory approval submissions.[14]
In both medical and pharma settings, while either making personalized treatment decisions or devising a clinical biomarker strategy, scientists and clinicians encounter biomarkers that may have a desired preclinical result or an altered readout in a patient, but have no qualified inference or validity linked to them. The probability of this is high as the majority of biomarkers are exploratory and only about 1% biomarkers are qualified. This is largely due to an active and unregulated biomarker discovery process and a more defined biomarker qualification process.
Biomarker discovery
The pace of biomarker discovery and validation has accelerated in recent years. New technologies, ground-breaking patient studies, and the completion of the human genome project have all contributed to a dramatic increase in the number of exploratory biomarkers. Fifty percent of all research articles referencing new cancer biomarkers have been submitted in the last decade alone [Figure 1].
Figure 1: Number of research publications appearing in PubMed for the term “cancer biomaker”
Biomarker qualification
Biomarker qualification is controlled by international medical regulatory bodies. The United States Food and Drug Association (FDA), in partnership with the European Medicines Agency (EMA), has developed a regulatory framework to qualify biomarkers as part of its Critical Path initiative. [1], [8] This framework ensures “Biomarker qualification is a graded, fit-for-purpose, evidentiary process linking a biomarker with biological and clinical endpoints [1], [12], [15], [16] required for a biomarker to be used as an endpoint in clinical trials that are intended to support the regulatory approval of a drug.”
Besides regulatory qualifications, biomarkers also qualify through inclusion into clinical practice guidelines based on real-world data evidence and in drug labels and as companion diagnostics for targeted therapies.
The gap between discovery and qualification
There is a pronounced imbalance between the number of biomarkers being discovered and those being qualified. Of the 40,000 recorded biomarkers, fewer than 400 are FDA-qualified for a specific context of use or listed in treatment guidelines. [2]–[5], [7], [8] Only 40 unique companion diagnostics are listed with the FDA. [6] This gap between exploratory and qualified biomarkers impacts the clinical transition of biomarkers for both improved drug trials and disease management. Any progress in bridging the gap would represent an opportunity in increasing the impact of biomarkers.
Given the volume of available biomarkers, the first step towards bridging the gap should be the ability to rationally prioritize biomarkers. Using large biomarker data sets such as Excelra’s Biomarker insights, researchers can compare all biomarkers using the validity index metric. This weighted multicomponent metric allows assessment of biomarkers standing within an indication based on readouts from multiple references. The biomarkers also remain associated with the measured clinical outcomes which are also quantified using the vast dataset. This ability to score and select biomarkers for a selected context of use helps improve trial design and increase the potential for eventual regulatory approval.
Developing a validity index and quantifying biomarker outcomes
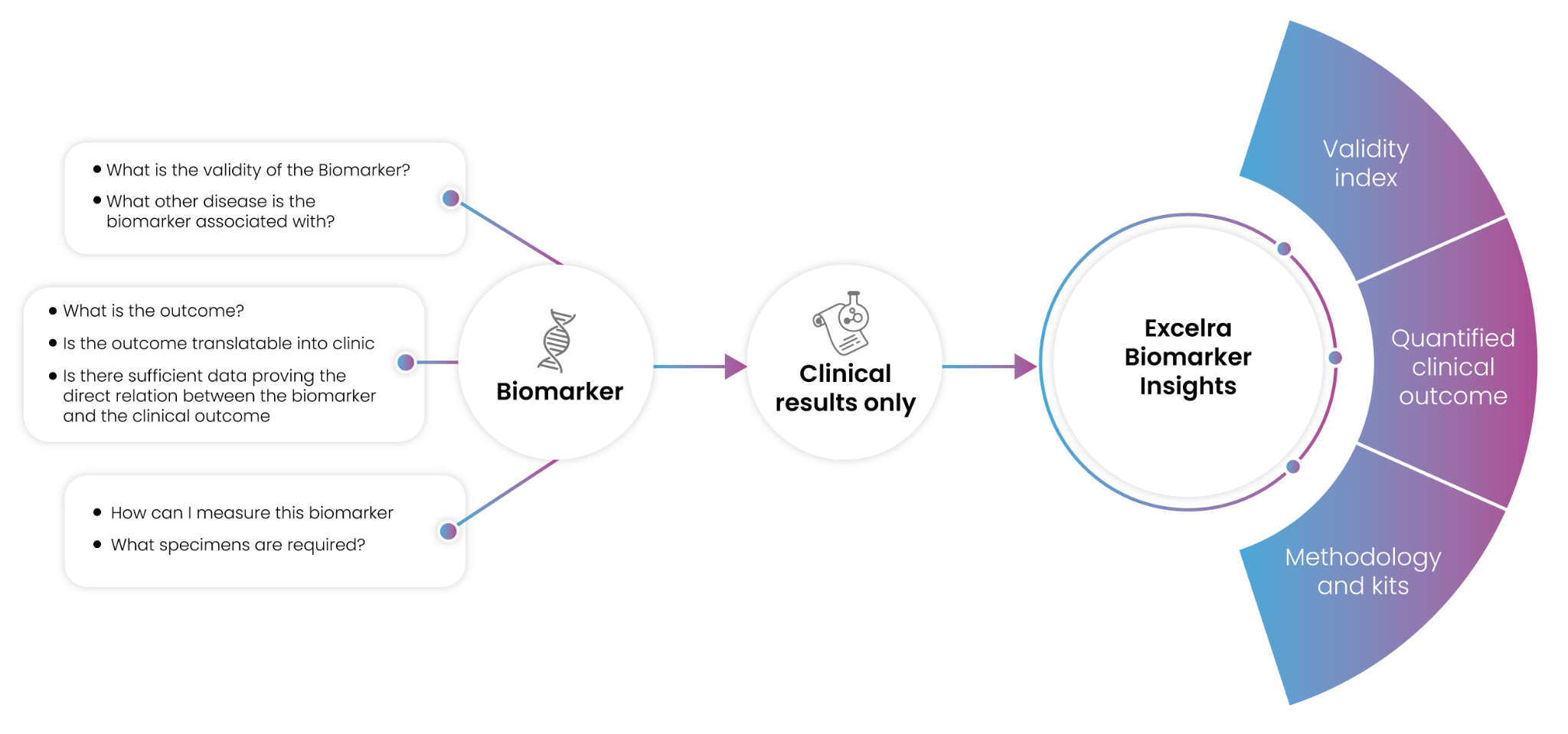
There are three primary aspects that can facilitate biomarker qualification: evidence of an association between biomarker and disease, evidence of an association between biomarker and a clinical outcome, and analytical verification of the biomarker in question. [Figure 2] Collecting a significant volume of evidence for biomarker-disease and biomarker-clinical outcome associations is challenging and requires building substantial curated datasets on which quantification of biomarker traits can be meaningfully applied.
Figure 2: Critical biomarker requirements (left) and solutions offered by Excelra’s Biomarker insights
Using a validity index to collect evidence of biomarker-disease association
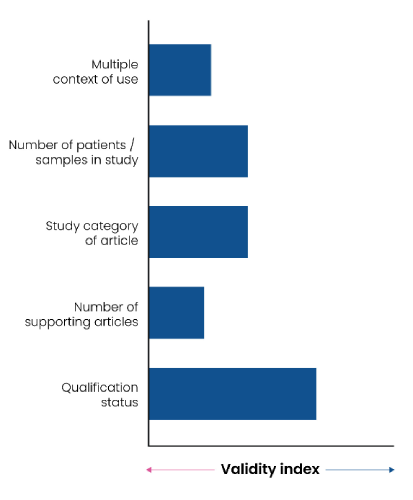
A biomarker’s context of use is a critical factor in its appraisal for qualification. A significant component of that context is the disease indication it is associated with. To quantify the strength of the association between biomarker and disease, researchers can use a validity index referencing multiple relevant factors. Excelra’s Biomarker insights ranks biomarkers according to a validity index comprising five factors. These factors are weighted according to their relative value in determining biomarker-disease association [Figure 3]:
-
Biomarker qualification status
Highest weightage is given to pre-existing regulatory qualification of the biomarker, its presence as a companion diagnostic, presence of the biomarker in any drug-label or clinical guidelines.
-
The number of supporting articles
This is a count of the number of distinct articles that provide evidence to the association of the biomarker to the disease.
-
The study category of those articles
This component rates the article category e.g., registered clinical trials get a higher rating than a case study.
-
The combined number of patients/samples referenced in the study
This component scores the number of patients recruited in a study e.g. a study with 500 samples would rank higher than a study with 50.
-
Multiple contexts of use for the biomarker
Multiple applications tagged with the biomarker in the disease will positively impact its scoring. For example, a biomarker with both predictive and prognostic applications will rank higher than a biomarker with only prognostic applications.
Quantifying evidence of biomarker-clinical outcome association
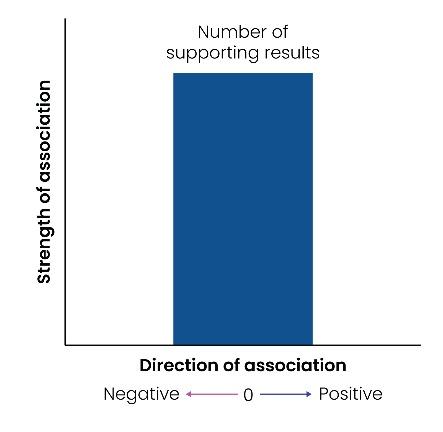
The clinical outcome associated with a biomarker gives granularity to its context of use. For example, a clinical outcome may further define the prognostic application of a biomarker by associating it with either metastasis, survival, or immune response. Excelra’s Biomarker insights quantifies the strength of this evidence according to the number of results associating the biomarker with the outcome. The score is represented as either negative or positive, depending on the specific result. For example, if increased biomarker level leads to metastasis it will return a positive score, and absence of metastasis will return a negative score [Figure 4]
Case study: validity and quantification results for biomarkers associated with colorectal cancer
To demonstrate the practical application of validity indices, we tested Biomarker insights validity index against biomarkers associated with colorectal cancer. There are 2,100 initial biomarker results for colorectal cancer on Biomarker insights. These biomarkers can be classified into subtypes based on variant, modification, specific long non-coding RNA (lncRNA), or microRNA (miRNA). With these additional subtypes, the total count reaches 3,700 results. Of these biomarkers, only 47 (1.3%) are qualified through their presence in drug labels, companion diagnostics, clinical guidelines, or approval for specific contexts of use.
Biomarker insights generated a validity index as described above and ranked the 3,700 biomarkers according to their scores. 31% of the resulting top 100 biomarkers were qualified biomarkers, endorsing this method of ranking [Figure 5]. Interestingly, while the highest-scoring biomarkers are qualified, several as-yet unqualified biomarkers also have a high validity index and can be used to reliably infer clinical implications. Table 1 includes further details on the top 25 biomarkers in the validity index, including their associated clinical outcomes, qualification status, and basis of their qualification.
Several unqualified biomarkers in the index were linked to a response to therapy application and associated with clinical outcomes such as overall survival, treatment response, immune cell infiltration, or prognostic applications with an association to metastasis and recurrence. The prioritization of these biomarkers would be beneficial in studies aimed to address unmet treatment needs originating from the inability to control metastasis or increase immune cell infiltration.
Figure 5: Top 100 colorectal cancer biomarkers based on validity score and qualification status
Table 1: List of top 25 colorectal cancer biomarkers as per their validity score. The number of articles, applications, clinical outcomes, and qualification basis, if any, have been listed.
| Sr. No. | Biomarker Name | Validity Score | Associated articles | Primary context of use application]- associated clinical outcomes (Positive association/Negative association) | Qualified biomarker [Y/N] and basis of qualification |
|---|---|---|---|---|---|
| 1 | CARCINOEMBRYONIC ANTIGEN (CEA) | 198.73 | 177 | [Diagnostic| Prognostic] Disease diagnosis | Disease stage/grade| Metastasis| Recurrence Disease-free survival| Overall survival |
[Y] CEA is FDA-approved cancer biomarker for monitoring colon cancer |
| 2 | KRAS VARIANT | 134.9 | 119 | [Prognostic| Response to therapy] Disease stage/grade |Metastasis| Treatment response Disease-free survival| Overall survival | Immune cell infiltration |
[Y] Presence of KRAS mutation is contraindicated for treatment with drugs like cetuximab, panitumumab, irinotecan, oxaliplatin-containing chemotherapy |
| 3 | BRAF VARIANT | 102.26 | 88 | [Prognostic| Response to therapy] Progression-free survival| Overall survival |
[Y] Prognostic assessment. RAF mutant tumors contraindicated for treatment with panitumumab or cetuximab |
| 4 | T CELLS | 94.73 | 96 | [Prognostic| Response to Therapy] Disease diagnosis| Overall survival| Treatment response Tumor invasiveness |
[N] |
| 5 | C-REACTIVE PROTEIN (CRP) | 73.87 | 67 | [Prognostic| Response to therapy] Organ damage/safety Overall survival |
[N] |
| 6 | CARBOHYDRATE ANTIGEN 19-9 (CA19-9) | 72.12 | 67 | [Prognostic| diagnostic] Disease diagnosis| Disease stage/grade Overall survival |
[N] |
| 7 | CD274 MOLECULE (CD274) | 71.05 | 67 | [Prognostic| Response to therapy] Disease diagnosis| Overall survival |
[N] |
| 8 | NEUTROPHIL TO LYMPHOCYTE RATIO (NLR) | 58.92 | 55 | [Prognostic| Response to therapy] Disease diagnosis| Disease stage/grade Disease-free survival | Overall survival |
[N] |
| 9 | CIRCULATING TUMOR DNA (ctDNA) | 57.69 | 52 | [Response to therapy] Recurrence Disease-free survival |
[Y] Circulating tumor DNA (ctDNA) may be used to complement multidisciplinary decision-making and is recommended to determine the optimal perioperative chemotherapy for CRC patients. |
| 10 | RAS VARIANT | 53.87 | 45 | [Prognostic| Predictive] Disease-free survival| Overall survival| Progression-free survival |
[Y] Cetuximab is indicated for patients with (EGFR)-expressing, RAS wild-type metastatic colorectal cancer; combination of cetuximab with oxaliplatin-containing contraindicated mutant RAS mCRC |
| 11 | INTERLEUKIN 6 (IL6) | 51.20 | 49 | [Prognostic| Response to therapy] Disease diagnosis |
[N] |
| 12 | EPIDERMAL GROWTH FACTOR RECEPTOR (EGFR) | 48.00 | 44 | [Prognostic| Response to therapy] Disease progression |
[Y] Cetuximab is indicated for patients with (EGFR)-expressing, RAS wild-type metastatic colorectal cancer |
| 13 | CD8+ T CELLS) | 44.65 | 44 | [Prognostic| Response to therapy] Immune cell infiltration| Overall survival| Treatment response Disease stage/grade| Tumor invasiveness |
[N] |
| 14 | TUMOR PROTEIN P53 (TP53) | 40.35 | 37 | [Prognostic| Response to therapy] Overall survival |
[N] |
| 15 | TUMOR NECROSIS FACTOR (TNF) | 39.23 | 38 | [Prognostic| Response to therapy] Disease diagnosis |
[N] |
| 16 | BRAF NP_4324.2:p.V600E | 39.12 | 27 | [Prognostic| Predictive] Overall survival| Progression-free survival |
[Y] Recommended treatment for BRAF V600E Mutation-Positive Metastatic Colorectal Cancer - combination therapy irinotecan/ cetuximab/ panitumumab/ vemurafenib/ encorafenib. |
| 17 | VASCULAR ENDOTHELIAL GROWTH FACTOR (VEGF) | 36.78 | 35 | [Diagnostic| Response to therapy] | [N] |
| 18 | MICROSATELLITE INSTABILITY (MSI) | 34.04 | 25 | [Prognostic] Immune evasion |
[Y] checkpoint inhibitors (pembrolizumab, nivolumab, ipilimumab) for dMMR/MSI high disease. |
| 19 | PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA (PIK3CA) | 33.5 | 32 | [Prognostic| Predictive[Prognostic| Response to therapy] Overall survival |
[N] |
| 20 | INTERLEUKIN 1 (IL1) | 33.18 | 33 | [Prognostic| Diagnostic] | [N] |
| 21 | ALBUMIN (ALB) | 32.1 | 29 | [Prognostic] | [N] |
| 22 | PLATELET TO LYMPHOCYTE RATIO (PLR) | 32.08 | 30 | [Prognostic| Response to therapy] Disease diagnosis Disease-free survival| Overall survival |
[N] |
| 23 | C-X-C MOTIF CHEMOKINE LIGAND 8 (CXCL8) | 31.87 | 29 | [Prognostic| Diagnostic] Disease diagnosis| Immune cell infiltration |
[N] |
| 24 | LYMPHOCYTES | 31.28 | 28 | [Prognostic] Disease-free survival| Overall survival |
[N] |
| 25 | APC REGULATOR OF WNT SIGNALING PATHWAY (APC) | 30.65 | 27 | [Prognostic| Diagnostic] Disease risk| Overall survival |
[N] |
Conclusion
The gulf between exploratory biomarkers and qualified biomarkers is vast: less than 1% of the biomarkers observed in clinical studies achieve regulatory qualification. This gap is likely to remain as long as researchers lack a standardized method of measuring biomarker validity.
One potential method is a validity index, as discussed here. With carefully selected criteria, all biomarkers- unqualified, exploratory as well as qualified, can be evaluated and ranked to aid prioritization, and the resulting index can reveal opportunities for further research. Further values are added by linking these biomarkers to quantified clinical outcomes.
Excelra’s Biomarker insights is a comprehensive database of manually curated validated and exploratory biomarkers, providing an overview of the biomarker-disease relationship. It contains rich data encompassing exploratory, pre-clinical, and clinical domains, and provides critical insights into diagnosis, prognosis, treatment response, safety, efficacy, and toxicity.
With its optimized validity index, Biomarker insights helps researchers quickly identify the most relevant biomarkers for further study, saving significant time and money. The insights available in Biomarker insights can improve trial designs, streamline project-critical decisions, and ultimately accelerate the translation of promising biomarkers into clinical practice.
To find out how Biomarker insights can accelerate your drug discovery and development programs, or to request a demo, please click here.
References
1. FDA-NIH Biomarker Working Group, BEST (Biomarkers, EndpointS, and other Tools) Resource. 2016. Accessed: Mar. 02, 2023. [Online]. Available: https://www.ncbi.nlm.nih.gov/books/NBK326791/
2. “GOBIOM: global online biomarker platform,” 2023. https://gobiomdbplus.com/ (accessed Mar. 16, 2023)
3. D. S. Wishart et al., “MarkerDB: an online database of molecular biomarkers,” Nucleic Acids Res, vol. 49, no. D1, pp. D1259–D1267, Jan. 2021, doi: 10.1093/nar/gkaa1067
4. US FDA, “Labelling Information| Drug Products,” 2022. https://www.fda.gov/drugs/development-resources/labeling-information-drug-products (accessed Mar. 15, 2023)
5. US FDA, “List of Pharmacogenomic biomarkers in drug labels,” 2023. https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling (accessed Mar. 15, 2023)
6. US FDA, “List of Approved Companion Diagnostic Devices,” 2023. https://www.fda.gov/medical-devices/in-vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-in-vitro-and-imaging-tools (accessed Mar. 15, 2023)
7. US FDA, “List of Qualified Biomarkers,” 2021. https://www.fda.gov/drugs/biomarker-qualification-program/list-qualified-biomarkers (accessed Mar. 15, 2023)
8. European Medicines Agency (EMA), “Opinion and letters of support on the qualification of novel methodologies for medicine development.,” 2023. https://www.ema.europa.eu/en/human-regulatory/research-development/scientific-advice-protocol-assistance/novel-methodologies-biomarkers/opinions-letters-support-qualification-novel-methodologies-medicine-development (accessed Mar. 15, 2023)
9. National Comprehensive Cancer Network, “NCCN- Guidelines,” 2023. nccn.org (accessed Apr. 05, 2023)
10. A. Hartmann, C. Hartmann, R. Secci, A. Hermann, G. Fuellen, and M. Walter, “Ranking Biomarkers of Aging by Citation Profiling and Effort Scoring,” Front Genet, vol. 12, May 2021, doi: 10.3389/fgene.2021.686320
11. M. Brunet et al., “Barcelona Consensus on Biomarker-Based Immunosuppressive Drugs Management in Solid Organ Transplantation,” Ther Drug Monit, vol. 38, no. Supplement 1, pp. S1–S20, Apr. 2016, doi: 10.1097/FTD.0000000000000287
12. Book: Rahbari R, Van-Niewaal J, and Bleavins MR, Biomarkers in Drug Discovery and Development. Wiley, 2020. doi: 10.1002/9781119187547
13. W. A. Colburn, “Biomarkers in Drug Discovery and Development: From Target Identification through Drug Marketing,” The Journal of Clinical Pharmacology, vol. 43, no. 4, pp. 329–341, Apr. 2003, doi: 10.1177/0091270003252480
14. M. Gromova, A. Vaggelas, G. Dallmann, and D. Seimetz, “Biomarkers: Opportunities and Challenges for Drug Development in the Current Regulatory Landscape,” Biomark Insights, vol. 15, p. 1177271920974652, Jan. 2020, doi: 10.1177/1177271920974652
15. J. A. Wagner, “Strategic approach to fit-for-purpose biomarkers in drug development.,” Annu Rev Pharmacol Toxicol, vol. 48, pp. 631–51, 2008, doi: 10.1146/annurev.pharmtox.48.113006.094611
16. C. Altar et al., “A Prototypical Process for Creating Evidentiary Standards for Biomarkers and Diagnostics,” Clin Pharmacol Ther, vol. 83, no. 2, pp. 368–371, Feb. 2008, doi: 10.1038/sj.clpt.6100451
How can we help you?
We speak life science data and help you unlock its potential.