Introduction
In the area of genomics and molecular biology, Next Generation Sequencing (NGS) epitomises an iconic shift. It is a cutting-edge technology that has transformed the landscape of genetic research, enabling unprecedented insights into the complexities of genomic information and rapidly sequencing RNA and DNA. This thorough blog aims to explain the complexities of NGS, from its central ideology to its vast uses in biomedical research study.
Introduced for commercial use in 2005, this method was initially called “massively parallel sequencing”, because it enabled the sequencing of many DNA strands at the same time, instead of one at a time as with traditional Sanger sequencing by capillary electrophoresis (CE) [1].
What is NGS and how it works?
The term “NGS, or Next Generation Sequencing”, refers to cutting-edge sequencing technologies that enable the parallel sequencing of countless minor DNA fragments. NGS can quickly and economically sequence whole genomes. This technological advancement offers an inclusive opinion of the genetic landscape, providing it as requisite for modern biomedical research.
NGS sequencing steps
The NGS process involves various thorough steps:
1. Library preparation:
This includes fragmenting the DNA and connecting adapters on both ends of the fragments. During sequencing, the adapters acts as binding locations for primers.
2. Clonal amplification:
To produce enough DNA for sequencing, the library of DNA is amplified. Bridge amplification or emulsion PCR are the techniques used to do this.
3. Sequencing of NGS library:
The nucleotide sequences of amplified library are sequenced by means of advanced platforms. Enormous volumes of data is generated in this process, which need to be related and analysed.
4. Data analysis of NGS:
To identify genetic variants, the raw sequencing data is analysed and processed. Interpretation of the biological importance of the sequenced genomes is contingent upon this stage.
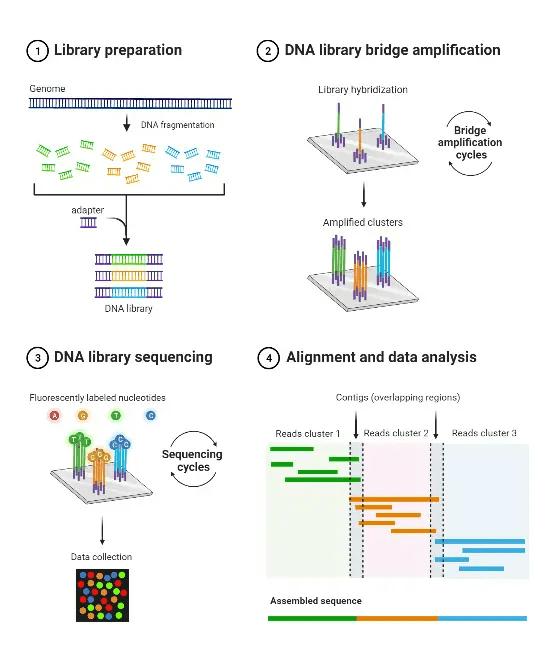
Figure 1: NGS Sequencing Steps
Types of next generation sequencing
Depending upon the research objective, NGS includes respective methodologies, each contributing to diverse advantages.
1. Whole genome sequencing (WGS):
WGS gives a broad view of the complete genome, letting researchers to learn genetic variants through the entire sequence of the DNA. This approach is principally beneficial for finding new mutations and deciphering intricate genetic diseases.
2. Whole exome sequencing (WES):
WES emphases on sequencing the genome’s exomic regions, which account for nearly 1-2% of the whole genome but also contains around 85% of identified disease-linked variants. This technique is very economical and highly enlightening for clinical uses.
3. Targeted sequencing:
This method involves sequencing area of interest within the genome. With high coverage and accuracy, it is idyllic for studies focussing on specific genes or genetic regions.
4. RNA sequencing (RNA-Seq):
RNA-Sequencing gives insights towards the transcriptome, illuminating patterns of gene expression and categorising new transcripts. In discovering biomarkers and understanding gene regulation, this method is contributory.
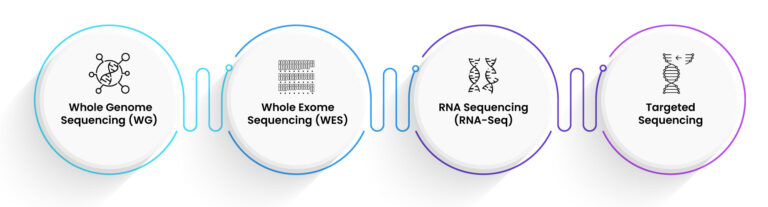
Figure 2: Types of Next Generation Sequencing
NGS applications in biomedical research
NGS technology has countless number of applications in the biomedical research, revolutionizing how scientists perform genetic research.
1. Drug discovery and development:
NGS makes it easy to identify the genetic targets for drug development, allowing personalized medicine methods. Researchers can create tailored therapies to improve outcomes of the patients by grasping the genetic root of the diseases.
2. Cancer genomics study:
NGS is utilized in oncology to identify genetic mutations that direct cancer advancement. This technology is crucial for invention of targeted cancer remedies, moreover, progressing diagnostic accurateness.
3. Research on infectious disease:
NGS is crucial in pathogen identification for infectious disease and tracing the dissemination of it. This is intended for rapid sequencing of viral as well as bacterial genomes, helping in outbreak management along with vaccine development.
4. Genetic disorder diagnosis:
For scarce genetic disorders, NGS offers an extensive diagnosis tool, permitting the identification of relevant mutations that might be skipped by traditional procedures.
Advantages of NGS
1. Higher coverage and sensitivity:
NGS provides a sensitive and thorough method than conventional Sanger sequencing, letting it for better identification of variations around the genome.
2. Faster sequencing:
NGS can sequence millions of DNA fragments concurrently, significantly dipping the time required for DNA analysis. It facilitates sequencing of whole genomes in 24 hours.
3. Cost-Effective:
The sequencing cost has radically reduced with NGS, making it more affordable and accessible for large-scale studies.
4. Ability to sequence anything:
NGS assent the sequencing of broad range of DNA samples, from specific target areas to whole genomes, advancing flexibility in their research.
5. Detection of genetic abnormalities:
NGS can identify a variety of genomic irregularities, including insertions, deletions, substitutions, duplications, and chromosomal inversions/ translocations, offering a thorough view of genetic variation.
6. Less DNA samples:
NGS requires much less DNA compared to traditional sequencing methods, enabling efficient use of limited samples.
7. Improvement for clinical applications:
For example, tumour specimens can be sequenced and analysed for genomic amendments within about 10 days, making it a applied tool in clinical diagnostics.
8. Comprehensive genome analysis:
With NGS, scientists can analyse the complete genome for abnormalities, smoothing more systematic genetic research, involving cancer genomics and personalized medicine.
Challenges in NGS data analysis
NGS data analysis reports numerous difficulties, particularly in the integration of varied data types like proteomics, genomics and transcriptomics. Conversely, modern advancements in bioinformatics have delivered robust solutions to all these obstacles.
1. Integration of data:
A comprehensive intelligence of biological systems requires the incorporation of multi-omics data. The accuracy of biological insights can be improved by facilitate the seamless integration of various data types made possible by computational tools and algorithms.
2. Data storage and management:
A huge data is generated from NGS which requires competent storing solutions. High-performance computing resources and cloud-based platforms are gradually increasing and utilized to oversee and analyse NGS data efficiently.
3. Interpretation of genetic variants:
Illuminating the functional importance of genetic variants continues as a challenge. Yet, software tools and databases are endlessly being remodelled to foster researchers prioritize and annotate variants for more research study.
Conclusion
Next Generation Sequencing is an essential technology that remains at centre of biomedical research innovation. Its capability to offer comprehensive genetic data has unlocked new possibilities for identifying complicated diseases and creating targeted therapies. As NGS technology progresses, it is a need for data analysts, researchers and science communicators to grab its difficulties and applications. By doing so, they can hitch the extensive ability of NGS to advance their relevant fields and add this to the wider scientific community.
Understanding NGS testing and its potential applications demands not only expertise with the technology but also ability in analysing and integrating the massive data it produces. This overview tends to the foundational source for those obtaining to increase their learning and utilization of NGS in the domain of biomedical research.
Since NGS is producing unprecedented amounts of data, the bottleneck has shifted from data generation to data processing & analysis. With its state-of-the-art bioinformatics pipelines, OP², Excelra’s Online Pipeline Platform, enables researchers to quickly gain actionable insights from massive amounts of data.
NGS Data Analysis by Excelra
Excelra delivers end-to-end genomics and transcriptomics data services designed to accelerate biomedical research, NGS data analysis, and drug discovery efforts. Central to this offering is the curation and interpretation of complex omics datasets, including RNA-Sequencing outputs, derived from diverse experimental models, clinical samples, and public repositories. The curated data is processed into standardized, analysis-ready formats, enabling consistent interpretation across tissues, conditions, and timepoints.
A cornerstone of Excelra’s capabilities lies in its Next-Generation Sequencing (NGS) Data Analysis, where raw sequencing data — including bulk RNA-seq, small RNA-seq, and single-cell data — is transformed into meaningful biological insights. Through advanced computational workflows and pipelines, Excelra identifies differentially expressed genes, signaling pathways, biomarkers, and regulatory elements that support target validation, mechanism-of-action studies, and disease pathway modeling.
Excelra also supports translational pipelines by integrating NGS data with other bioinformatics and cheminformatics tools. This synergy enables researchers to correlate molecular signatures with compound responses, bridging gaps between discovery biology and therapeutic development. These capabilities are particularly critical for early-stage drug discovery programs, where insights into gene expression profiles inform target prioritization and lead optimization.
An added differentiator is Excelra’s use of AI/ML algorithms and proprietary ontologies to scale insight generation. This includes automated annotation of gene functions, clustering of transcriptomic patterns, and creation of molecular interaction networks. Such approaches are invaluable for large-scale studies involving patient cohorts or high-throughput screening data.
Through its genomics offerings, Excelra enables research institutions and life sciences companies to move from fragmented data to integrated insights. Whether applied to disease mechanism studies, biomarker discovery, or clinical translational research, Excelra’s solutions empower a data-driven approach to biomedical innovation.

