Overview
This case study outlines Excelra’s data-driven approach to performing tumor-enriched target identification, prioritization, and lineage-specific biomarker discovery for small cell lung cancer (SCLC) and ovary carcinoma (OC). The work involved public oncology research data from TCGA and GEO, bulk and scRNA-seq analysis, and multi-database gene annotation—enabling actionable insights for precision oncology efforts.

Our client
A pharma company based in the EU, actively working in antiviral development, expanded its R&D focus into oncology research. They partnered with Excelra to utilize our domain-specific bioinformatics pipeline and expertise in data curation for tumor and lineage-specific target identification.

Client’s challenge
The client needed to collect and integrate bulk RNA and scRNA-seq datasets for SCLC and ovary cancer, along with vital normal organ controls. Their challenge involved:
- Accessing curated TCGA and GEO cancer datasets
- Performing comparative expression profiling between cancer and control groups
- Identifying tissue-restricted and lineage-specific targets with high therapeutic potential
- Structuring data with relevant gene annotation for clinical and translational use
These tasks demanded an efficient, reproducible, and scalable bioinformatics pipeline that could work across heterogeneous sources.

Client’s goals
- Perform tumor-enriched and lineage-specific target identification
- Integrate bulk and scRNA expression datasets with data curation and normalization
- Apply gene annotation frameworks to assess target druggability and clinical relevance
- Support downstream research with annotated gene lists, clinical pathways, and therapeutic links for precision oncology decision-making
Our approach
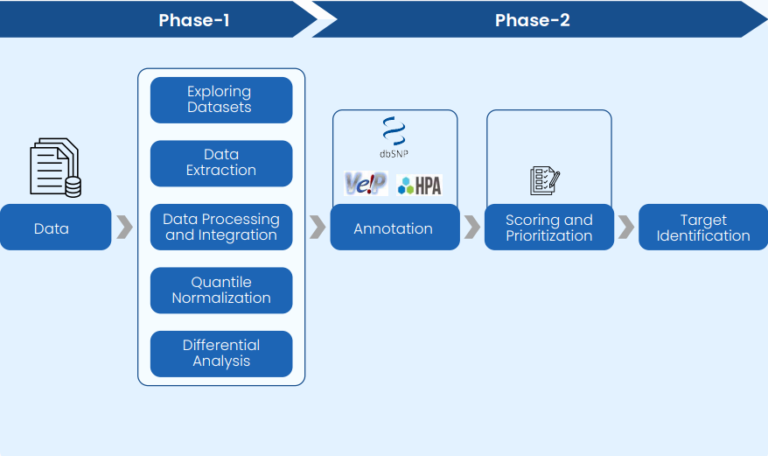
Excelra executed a two-phase approach, enabled by its proprietary bioinformatics pipeline and oncology research expertise:
Phase 1: Data Curation and Processing
- Retrieved high-quality bulk RNA and scRNA datasets from TCGA and GEO
- Conducted data curation and integration using robust QC and metadata checks
- Applied batch effect removal and quantile normalization to reduce variance
Phase 2: Gene Analysis and Prioritization
- Conducted differential expression analysis across tumor and control datasets
- Identified upregulated genes specific to SCLC and OC
- Refined findings using lineage-specific expression profiles from vital normal organs
- Applied multi-source gene annotation (pathways, clinical connections, druggability)
Prioritized genes based on scoring metrics relevant to precision oncology
Data Curation & Integration
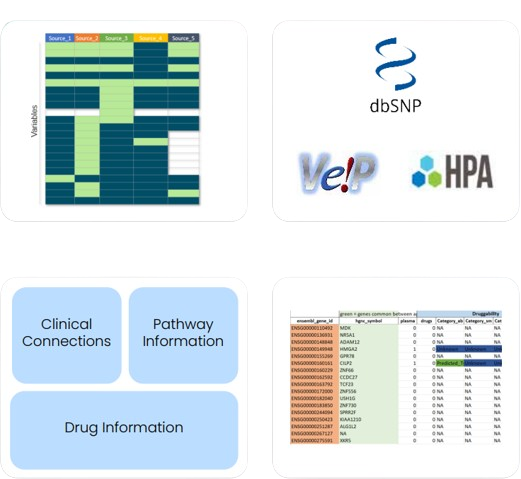
As part of Excelra’s oncology research initiatives, publicly available datasets from TCGA and GEO were collected for small cell lung carcinoma (SCLC), ovary carcinoma (OC), and their respective normal tissue controls. scRNA-seq data for tumor and normal tissues (lung, ovary, and other vital organs) were also integrated using our advanced bioinformatics pipeline and expert-led data curation process.
Normalization & Batch Correction
Quantile normalization and batch effect removal were applied across datasets to ensure uniformity and reduce technical variance. These steps were essential to maintain high-quality inputs for precision oncology analytics.
Target Identification
- Tumor Enrichment Analysis revealed differentially upregulated genes specific to SCLC and OC compared to their respective normal tissues.
- Lineage-Specific Target Analysis was performed by comparing cancer datasets with vital normal organ data to filter out broadly expressed genes using scalable bioinformatics pipelines.
- scRNA-based Analysis allowed precise identification of tumor-restricted genes for SCLC and OC by comparing expression at single-cell resolution against lineage-defining normal cells.
Annotation & Prioritization
Identified genes were annotated using multiple biological and pharmacological databases. This gene annotation process enabled robust target prioritization based on expression specificity, pathway involvement, druggability, and clinical relevance in the context of precision oncology.
Deliverables Generated
- A curated list of tumor-enriched and lineage-restricted targets for SCLC and OC, structured using Excelra’s proprietary data curation framework
- Prioritized gene sets for further preclinical validation aligned with oncology research goals
- Detailed gene annotation including expression profile, known drugs (if any), and disease relevance
- Final report and structured data files for downstream use in precision oncology pipelines
Conclusion
By leveraging bulk and scRNA expression analysis with structured bioinformatics pipelines, Excelra delivered a curated and prioritized gene list with clear tumor and lineage specificity. This included:
- Tumor-enriched gene buckets
- Lineage-restricted targets
- Gene annotation linked to clinical data, pathways, and pharmacological information
- Deliverables structured for downstream R&D and translational studies in precision oncology
Excelra’s integrated approach exemplifies how scalable data curation, high-confidence bioinformatics pipelines, and expert oncology research can drive innovation in cancer target discovery.

