Overview
Excelra partnered with a biotech innovator to develop a scalable, automated pipeline for high-quality bat genome assembly. Utilizing PacBio HiFi and Hi-C sequencing data, the pipeline achieved >98% genome completeness and >99.9% base-level accuracy. It integrated long-read and Hi-C data for chromosome-scale assemblies and was modularly designed to support viral genome assembly as well. The solution reduced manual effort by 75% and accelerated early discovery and therapeutic target identification. With validation-ready outputs and cross-platform compatibility, Excelra’s framework empowered the client to scale genomic research efficiently, enhancing their competitive edge in bat immunogenomics.

Our client
A biotech innovator at the forefront of bat genome research and studying their evolutionary adaptations. By leveraging the knowledge of bat genomics, the client was working towards developing therapeutic strategies, aiming to uncover unique biological traits and immune pathways in bats. The client was in early discovery stages and needed technical acceleration through bioinformatics automation. To achieve this goal, the client wanted Excelra to provide a framework for automating the assembly of PacBio HiFi sequencing reads plus Hi-C data to construct reliable bat genome assemblies.

Client’s challenge
An early-stage biotech company focused on studying bats’ evolutionary adaptations had generated large amount of data from sequencing of bat genomes. Bats are one of the most diverse mammalian orders, with over 1,400 species. Their genomes exhibited significant divergence, making comparative genomics and reference-guided assembly more difficult. To overcome this, researchers had to rely on high-quality de novo genome assembly using long-read sequencing technologies, to accurately capture both shared and species-specific genomic features.

Client’s goals
Wanted to build an efficient high-quality workflow for automating the assembly process of reference genome assemblies from scratch. They required a robust, automated pipeline to:
- Assemble PacBio HiFi reads
- Integrate Hi-C data for scaffolding
- Enable consistent and scalable genome assembly workflows
- Extend the framework to support virus genome assemblies from Illumina RNA-Seq data
Our approach
Excelra designed and deployed an end-to-end genome assembly pipeline tailored to high-accuracy PacBio HiFi sequencing data and Hi-C scaffolding. Additionally, we supported viral genome assembly from short-read data. The initial objective of the engagement was to standardize de novo assembly pipeline for Bat genomes using various sequencing technologies (PacBio, 10X, Bionano and Hi-C)
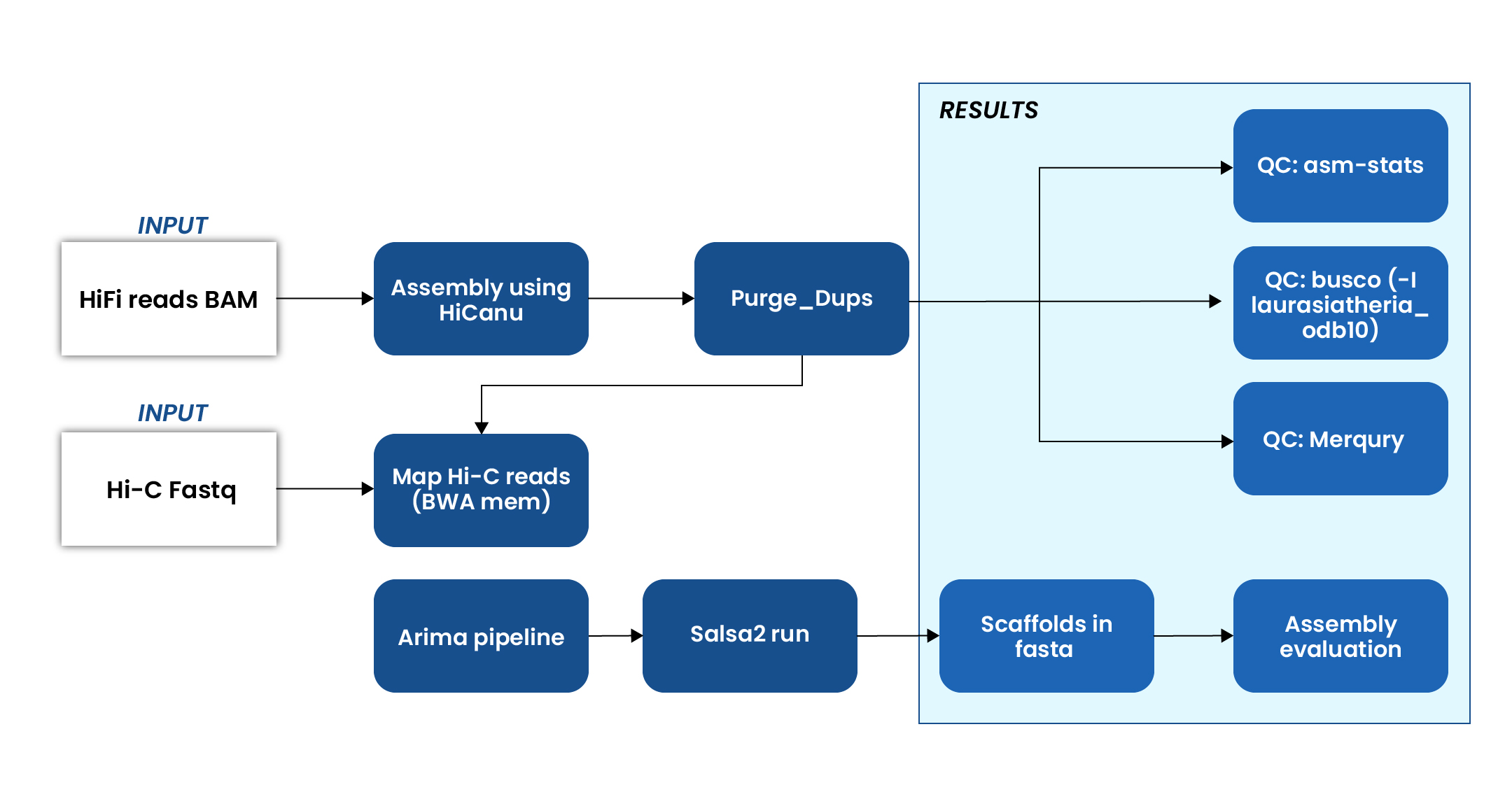
Genome Assembly Workflow
Input data
- PacBio HiFi BAM files for high-accuracy long-read sequencing.
- Hi-C fastq files for capturing chromatin interaction patterns.
HiFi assembly:
- Performed initial genome assembly using a tool, optimized for PacBio HiFi reads.
- Cleaned the assembly to remove haplotypic duplications and improve contiguity.
Hi-C integration
- Aligned Hi-C reads to the draft assembly using a tool for accurate mapping.
- Processed the mapped Hi-C data to prepare scaffolding input.
- Scaffolded the assembly using a tool, which utilizes Hi-C contact data to order and orient contigs into chromosome-scale scaffolds.
Assembly quality assessment
Conducted a comprehensive evaluation of the genome assembly using multiple tools to ensure accuracy, completeness, and structural integrity:
- Generated key assembly statistics such as N50, total assembly length, and number of contigs/scaffolds, providing a snapshot of assembly contiguity.
- BUSCO (Benchmarking Universal Single-Copy Orthologs) analysis was performed using the laurasiatheria_odb10 lineage dataset to assess the completeness of the assembly in terms of conserved gene content.
- Estimated base-level accuracy and completeness using a k-mer-based approach, comparing the assembly to raw sequencing reads for validation.
Final outputs
- A scaffolded genome in FASTA format, representing the assembled and Hi-C–scaffolded genome sequence at chromosome-level resolution.
- K-mer multiplicity plots, used to visualize and validate the consistency between the raw read data and the final assembly, helping to identify potential errors, duplications, or missing regions.
Our solution
Delivered a modular, automated pipeline for de novo assembly of bat genomes using PacBio HiFi reads and Illumina Hi-C reads , enabling:
- High-quality, reproducible reference genomes
- Smooth integration of long-read and Hi-C data
- Validation-ready outputs for downstream research. The assembly metrics, reads k-mer validation, and orthologous genes validation show that we were able to assemble a correct genome from HiFi and Hi-C reads.
Key performance outcomes
>98% assembly completeness, confirmed the presence of most conserved genes in the assembled genome.
>99.9% base-level accuracy, ensured high fidelity of the assembly with minimal sequencing or structural errors.
Reduced manual effort and turnaround time by 75%, enabling rapid generation of reference-quality genomes for multiple bat species.
Cross-platform data integration, reduced integration complexity and data processing costs.
Modular design for reusability, reproducible and scalable genome assembly workflows for future species or projects, eliminating the need to re-engineer pipelines.

Strategic business impacts
- Accelerated early discovery pipeline: High-quality assemblies enabled faster comparative and functional genomics research.
- Enhanced therapeutic target discovery: Accurate genomes unlocked unique biological traits for novel therapeutic target identification.
- Future-ready infrastructure: Modular pipeline supports future genome assemblies for bats and related pathogens.
- Improved data confidence for downstream analysis: Validation-ready outputs boosted confidence in downstream analyses with minimal additional processing.
- Strengthened competitive advantage: Early access to high-fidelity genomes positioned the client ahead in bat immunogenomics research.
Conclusion
Excelra developed and delivered a scalable, automated genome assembly pipeline that enabled a biotech client to generate high-quality, chromosome-scale bat genome assemblies by integrating PacBio HiFi and Hi-C sequencing data. The pipeline achieved >98% assembly completeness and >99.9% base-level accuracy, while reducing manual effort and turnaround time by 75%. Its modular, cross-platform design ensured seamless integration of diverse data types and positioned the client to rapidly scale future genome projects without re-engineering. This solution not only accelerated the client’s early discovery pipeline and enhanced therapeutic target identification but also provided validation-ready outputs that improved data confidence and strengthened their competitive edge in bat immunogenomics research.

