A brief review of GPCR family: The largest family of druggable targets
G protein-coupled receptors (GPCRs) have become a hot frontier in basic research of life sciences and therapeutic discovery of translational medicines and is widely pursued by both academic and industrial research for drug discovery. They represent an important opportunity for both small molecule-based and antibody-based therapeutics and are the largest family of targets for approved drugs. The discovery of a diverse set of molecules targeting this family could become valuable assets, by solving unexploited horizons like establishing target biological functions and disease relevance.
GPCR structures and families
GPCRs are the largest family of proteins involved in membrane signal transduction and are also the most intensively studied drug targets, largely due to their substantial involvement in human pathophysiology. The pharmacological modulation of GPCRs provides leverage for treatment of diseases of central nervous system (CNS), cancer, viral infections, inflammatory disorders, metabolic disorders, etc.
The superfamily is classified into six classes based on amino acid sequence similarities namely, Class A (rhodopsin-like family); Class B (secretin receptor family); Class C (glutamate receptor family); Class D (fungal mating pheromone receptors); Class E (cAMP receptors) and Class F (frizzled or smoothened receptors), of which only four (A, B, C and F) are found in humans.
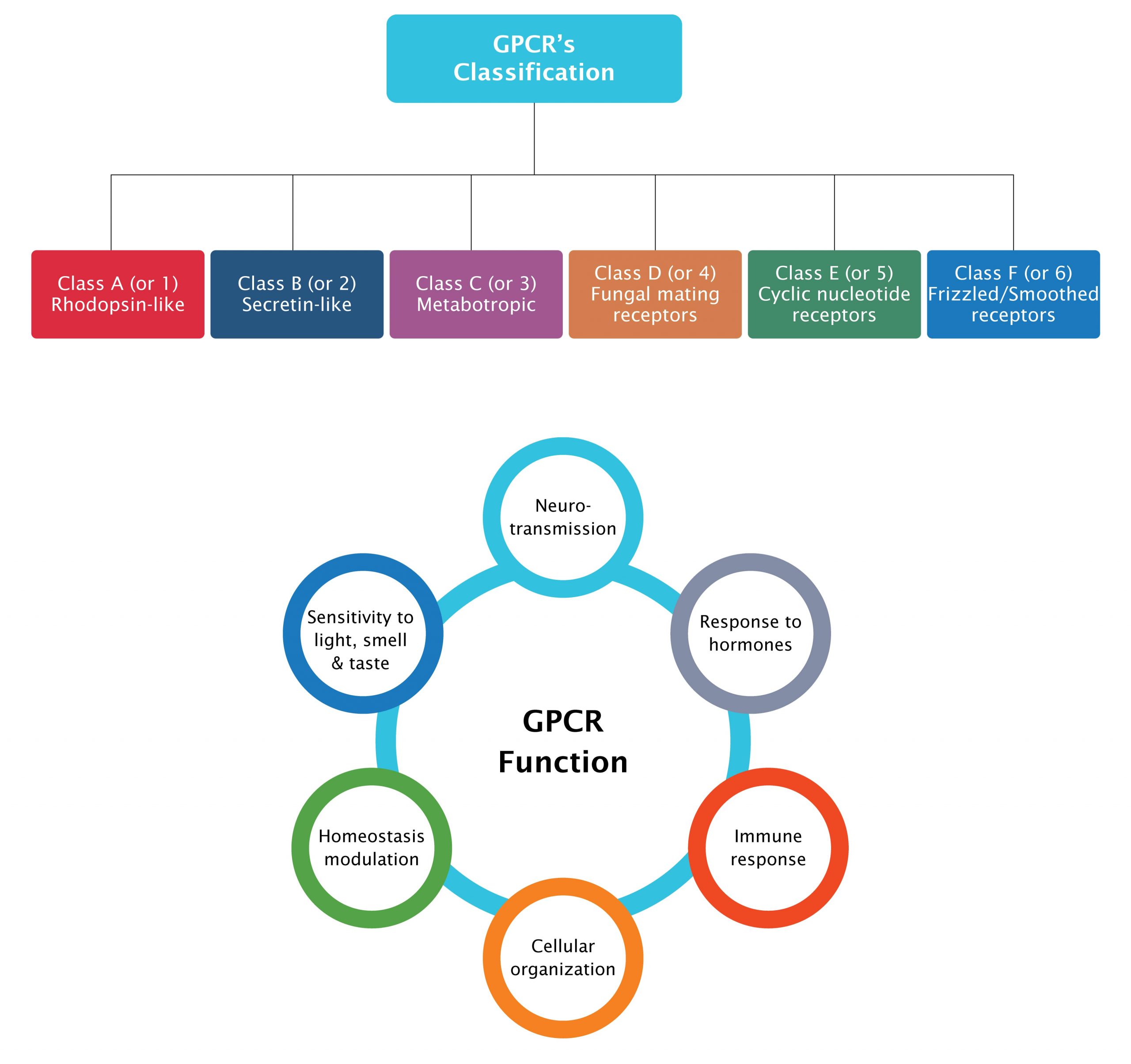
Figure 1: Target classes in GPCR super family
GPCRs are involved in various biological processes and disease indications and they make excellent drug targets (Fig 2). Some GPCRs have been linked to cancer development and progression, based on their overexpression and/or up-regulation by diverse factors. A higher expression of GPR49 was found to be involved in the formation and proliferation of basal cell carcinoma, the glycine receptor GPR18 was found to be associated with melanoma metastases, and high levels of GPR87 were found to be associated with lung, cervix, skin, urinary bladder, testis, head and neck squamous cell carcinomas.
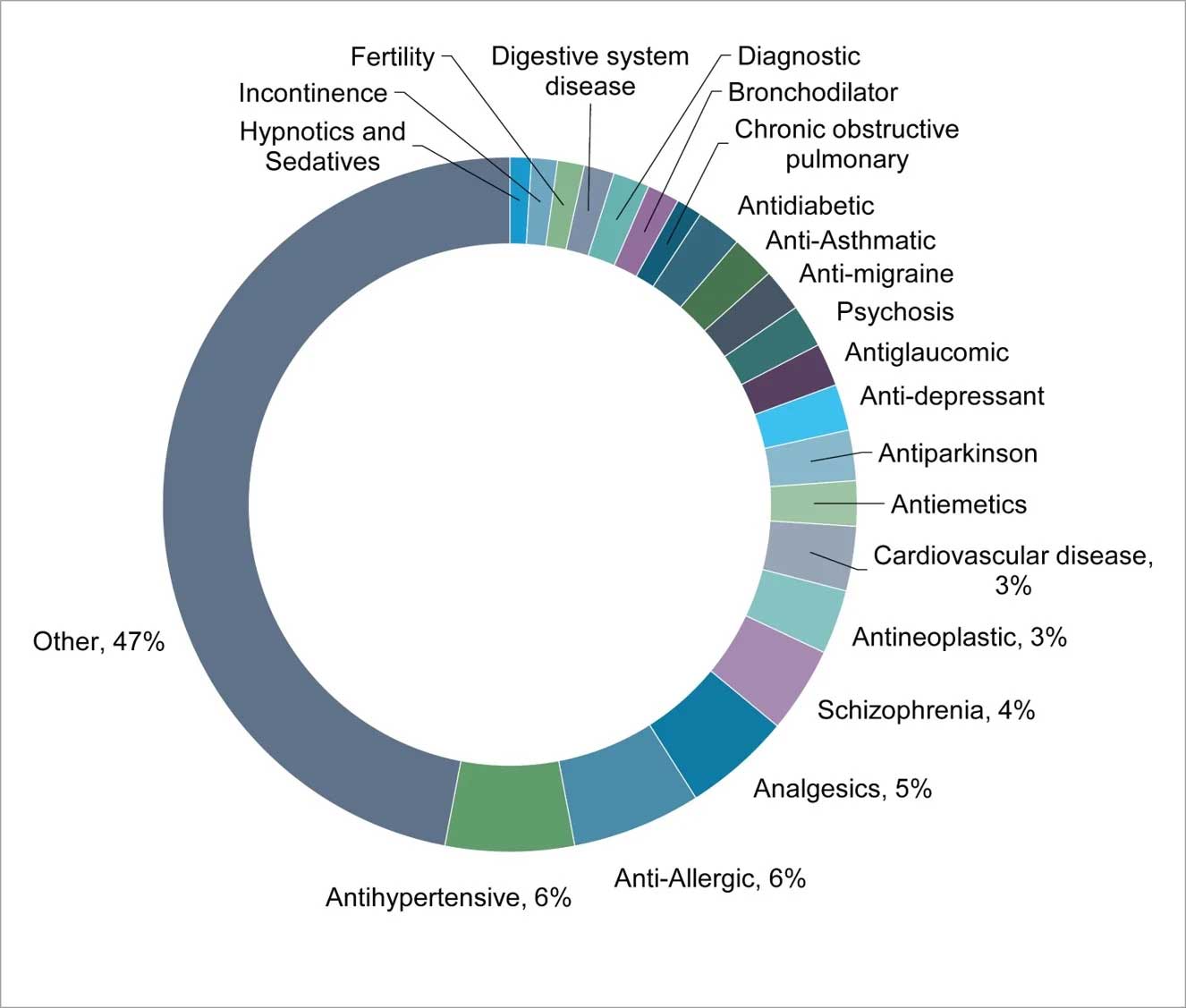
Figure 2: Some of the indications linked to members of GPCR family
Recently, orphan GPCRs have become a potentially novel targets for treatment of diverse set of indications, such as GPR119 for treatment of diabetes, leucine-rich repeat-containing G protein-coupled receptors 4 & 5 (LGR4/5) for treatment of gastrointestinal disease, GPR35 for treatment of an allergic inflammatory condition, GPR55 as an antispasmodic target, proto-oncogene Mas for treatment of thrombocytopenia, and GPR84 for of ulcerative colitis.
Landscape of GPCR research and drug development
GPCRs are the largest ‘target’ class of the ‘druggable genome’ representing approximately 19% of the currently available drug targets. In humans, the GPCR superfamily consists of 827 distinct members, of which 406 are non-olfactory. However, current therapeutics in humans target only 25% of potentially druggable GPCRs, 103 out of possible 403 GPCR targets, for which there is at least one marketed drug in practice.
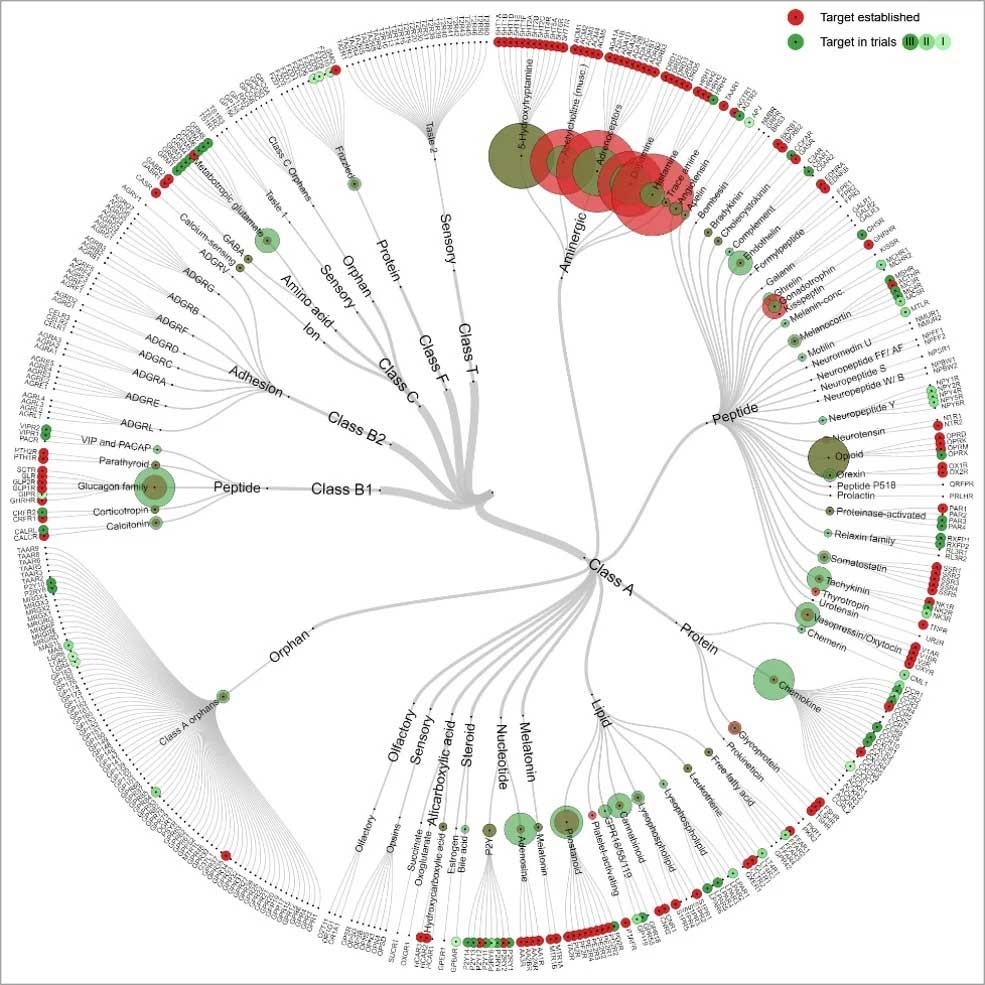
Figure 3: Classification tree of drugs targeting GPCR family members
Current literature analysis shows that GPCRs have traditionally been regarded as the domain for small-molecule drugs and very few targets are well studied. More than 30% of the US Food and Drug Administration (FDA) approved drugs target GPCRs, which makes them the largest druggable class of biomolecules (Fig 4).
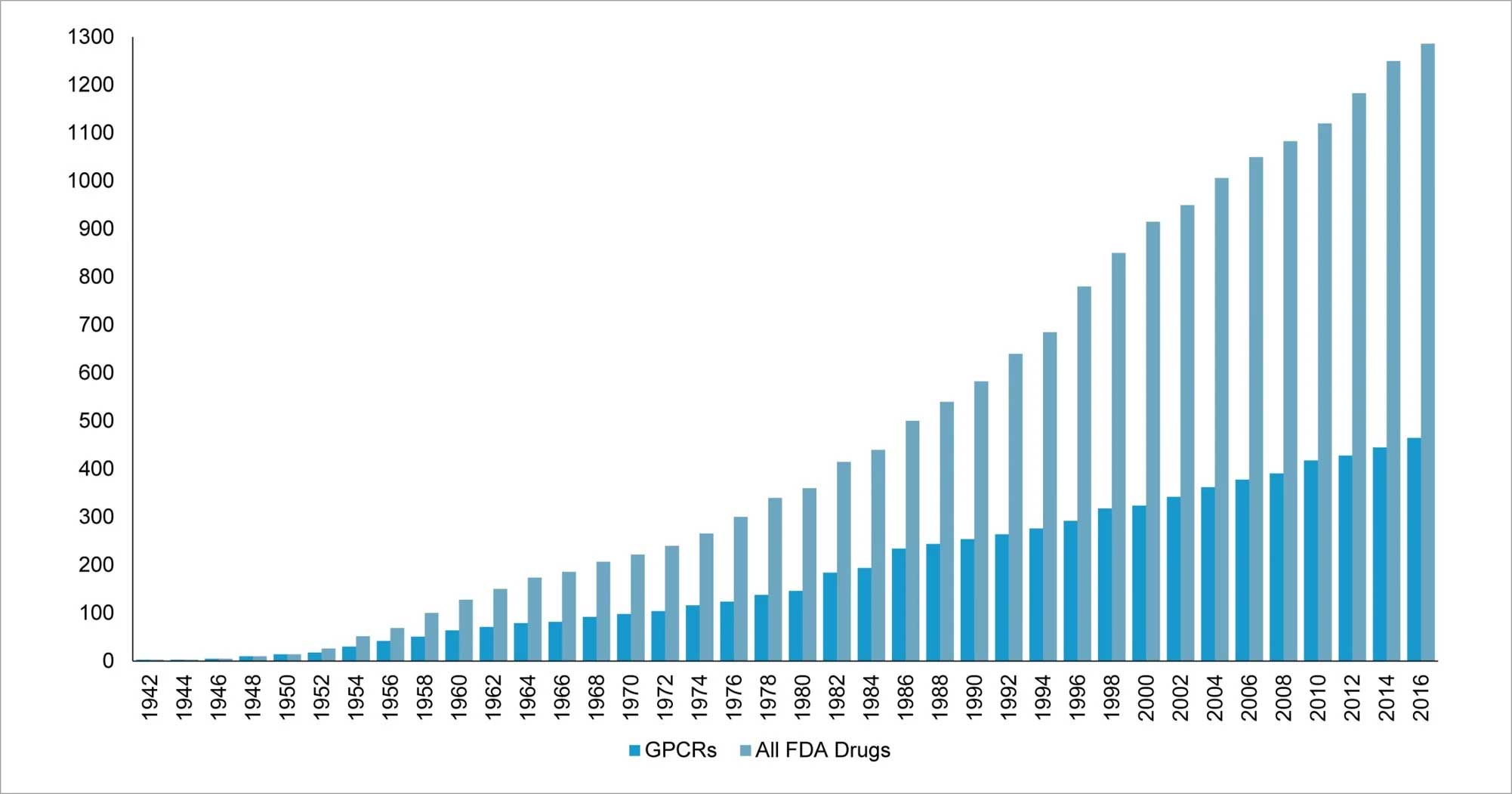
Figure 4: More than 30% FDA approved drugs target members of GPCR family
Enormous efforts have been expended to find relevant and potent GPCR ligands as lead compounds. Non-olfactory GPCRs constitute more than half of the human genome encoded targets that are not yet exploited for any therapeutic use and the knowledge is disproportionately focused in the scientific literature. Preliminary studies highlight that these receptors have functions in genetic and immune system disorders.
While the drugs that currently target GPCRs are primarily small molecules and peptides, GPCRs also recognize diverse ligands, including inorganic ions, amino acids, proteins, steroids, lipids, nucleosides, nucleotides, and small molecules (Fig 5).
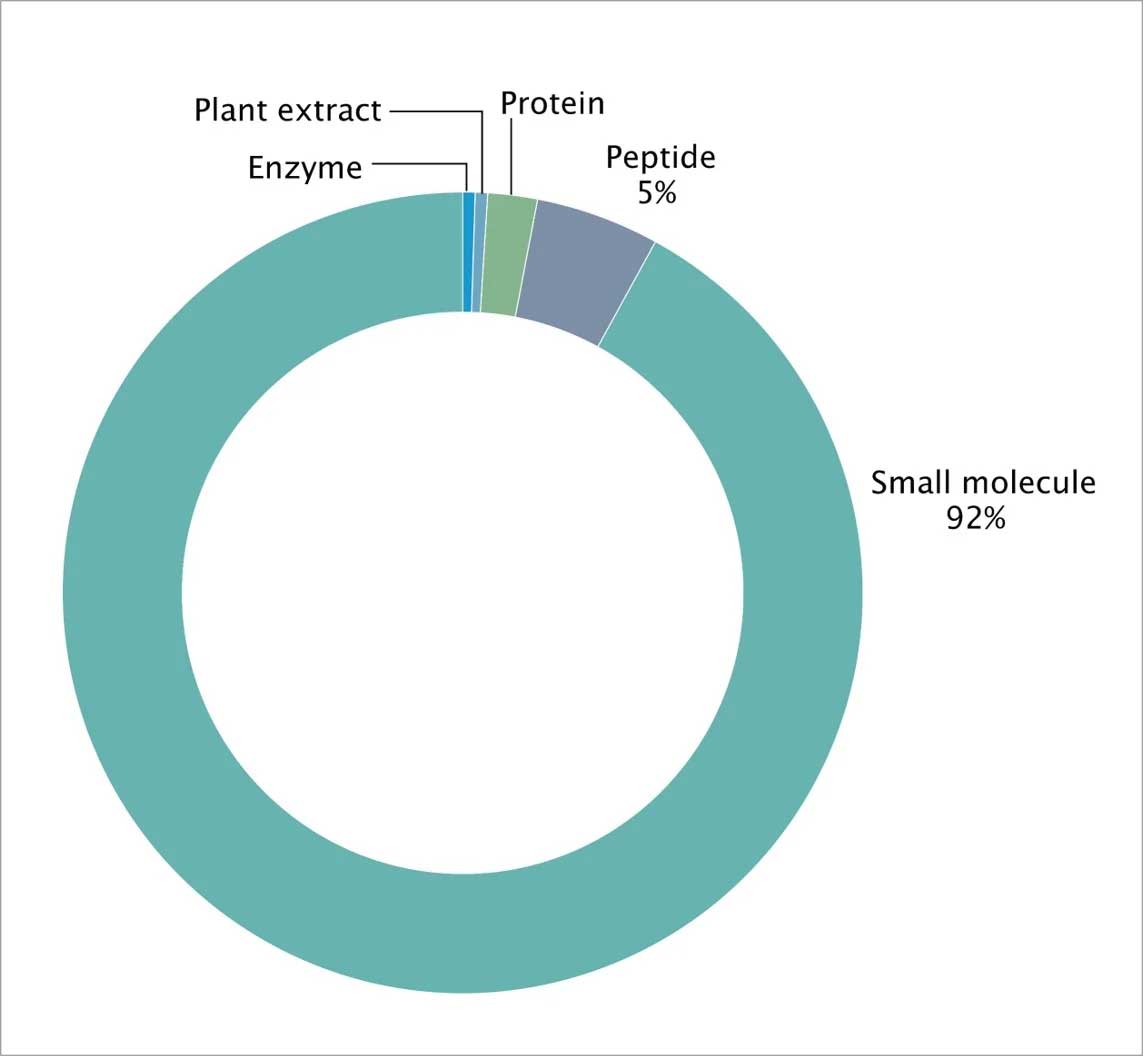
Figure 5: FDA approved drug types across GPCR classes
The latest trends in GPCR research indicates that modalities other than small molecules are becoming more popular as GPCR targeting agents with the entry of monoclonal antibodies, peptide drugs and allosteric modulators into early-stage clinical trials. For instance, GLP1 receptor targeting biologics like exenatide, liraglutide, and dulaglutide have been approved for type 2 diabetes, and CGRP receptor targeting erenumab in the treatment of chronic migraine and so many other peptide drugs targeting various GPCRs are also in development.
Current trends in GPCR research
In recent years, there is a significant increase in information available about the sequences, structures and signaling networks of GPCRs and the G proteins, due to breakthroughs in X-ray crystallography and cryo-electron microscopy (cryo-EM), leading to great understanding of GPCR-G protein interactions. This significant increase in information of GPCR-G protein interactions is being explored using several bioinformatics and software tools, including protein data bank , GPCRdb , gpDB , human gpDB and many more.
Due to limited spatial and high cost of experimental studies, computational modeling techniques such as bioinformatics, protein-protein docking and molecular dynamics simulations are playing an important role in exploring the GPCR-G protein interactions. Determining the 3-dimensional structural features of various unexplored orphan receptors and their ligand-associated complexes has become an exciting avenue in the GPCRs research in understanding on the molecular recognition and activation mechanisms and help the pharmaceutical investigation of new diseases in variety of therapeutic areas.
As the current human therapeutics cover only 25% of potentially druggable GPCRs, a relatively large extent of GPCRs still remain ‘orphan’ and therapeutically unexploited. This prediction and identification of GPCR ligands for these orphan receptors is an active area of research and interest to pharmaceutical industry.
References
- Hutchings CJ. A review of antibody-based therapeutics targeting G protein-coupled receptors: an update. Expert Opin Biol Ther. 2020 Aug;20(8):925-935
- Ellaithy A, Gonzalez-Maeso J, Logothetis DA, Levitz J. Structural and Biophysical Mechanisms of Class C G Protein-Coupled Receptor Function. Trends Biochem Sci. 2020 Dec;45(12):1049-1064
- Sriram K, Insel PA. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol. 2018 Apr;93(4):251-258
- Hauser AS, Attwood MA, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017 Dec;16(12):829-842
- Rask-Andersen M, Masuram S, Schioth HB. The druggable genome: evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu Rev Pharmacol Toxicol. 2014;54:9-26.
- Lu S, Zhang J. Small molecule allosteric modulators of G-protein-coupled receptors: drug-target interactions. J Med Chem. 2018
- Gugger M, White R, Song S, Waser B, Cescato R, Rivière P. GPR87 is an overexpressed G-protein coupled receptor in squamous cell carcinoma of the lung. Reubi JC Dis Markers. 2008; 24(1):41-50
- M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne. The Protein Data Bank Nucleic Acids Research, 2000;28:235-242
- Margarita C Theodoropoulou, Pantelis G Bagos, Ioannis C Spyropoulos and Stavros J Hamodrakas. “gpDB: A database of GPCRs, G-proteins, Effectors and their interactions.” Bioinformatics. 2008 Jun 15;24(12);1471-2
- Satagopam, V.P., Theodoropoulou, M.C., Stampolakis, C.K., Pavlopoulos, G.A., Papandreou, N.C., Bagos, P.G., Schneider, R. & Hamodrakas, S.J. GPCRs, G-proteins, effectors and their interactions: human-gpDB, a database employing visualization tools and data integration techniques. Database (Oxford) 2010;baq019
- Kooistra AJ, Mordalski S, Pándy-Szekeres G, Esguerra M, Mamyrbekov A, Munk C, Keserű GM, Gloriam DE. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Research, 2020;49:D335-D343


