Overview
Antisense Oligonucleotides (ASOs) are short, chemically modified strands of nucleotides designed to bind to complementary RNA sequences, enabling modulation of gene expression by altering splicing, blocking translation, or promoting transcript degradation. Among the different classes, RNase H-dependent ASOs recruit RNase H, an endogenous enzyme that cleaves the RNA strand of an RNA-DNA duplex. Unlike splice-switching or steric-blocking ASOs, these induce RNA degradation, offering unique therapeutic advantages.
RNase H-dependent ASOs are particularly versatile compared to RNA interference approaches such as siRNA due to their activity in both the nucleus and cytoplasm, fewer sequence design constraints, and flexible delivery methods. These features make them a promising modality in RNA therapeutics, with applications across neuromuscular, cardiovascular, metabolic, liver, and rare genetic disorders.
However, ASOs can also bind transcripts with partial complementarity, leading to unintended off-target effects that may cause pharmacological or toxicological consequences. Reliable detection, RNA-seq analysis, and characterization of these interactions are therefore essential during ASO design, safety assessment, and drug discovery pipelines. This case study highlights how Excelra’s scientific informatics expertise and machine learning accelerated ASO discovery by refining off-target prediction frameworks and significantly expanding transcriptome alignment coverage.
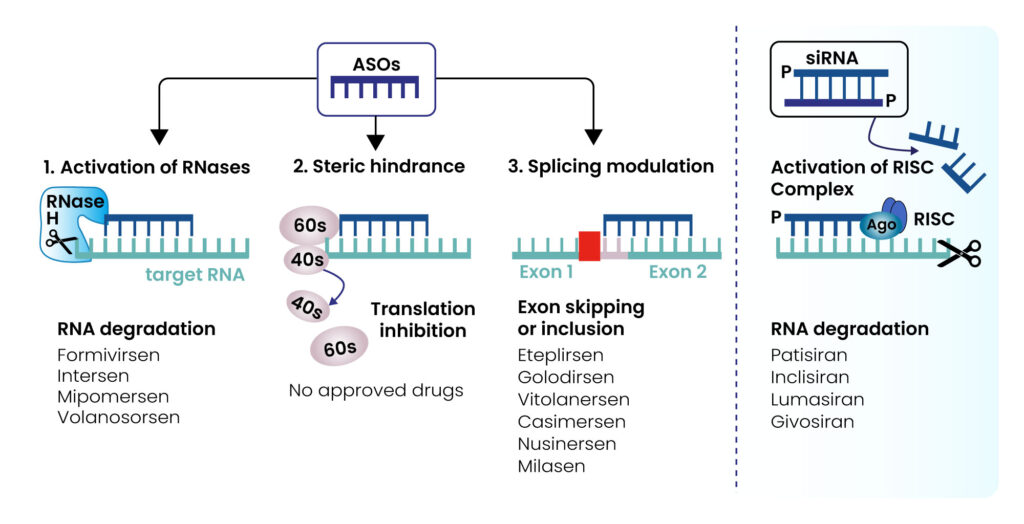
Credit Figure: Collota D. et al. 2023

Our client
The client, a global pharmaceutical innovator, was focused on advancing RNA therapeutics through antisense oligonucleotide (ASO) research and development. They sought expertise in bioinformatics and data-driven discovery to enhance their ASO screening process, particularly around precise off-target detection and interpretability of predictive models.

Client’s challenge
Despite having an internal machine learning model, the client faced critical limitations that slowed their RNA therapeutic pipeline:
- Rigid rule-based definitions: Existing off-target identification relied on basic sequence complementarity rules, leading to high false positives and missing biologically relevant interactions.
- Limited interpretability of ML models: Their black-box predictive model lacked transparency, reducing scientific trust and adoption by experimental teams.
- Constraints in sequence alignment: Fixed mismatch thresholds restricted transcriptome scanning and prevented detection of unconventional binding patterns.
- Lack of validation framework: No systematic method existed to evaluate or refine design rules based on empirical RNA-seq data.
These challenges necessitated a scalable, interpretable, and data-driven approach to enhance ASO discovery and improve the robustness of off-target prediction.

Client’s goals
The primary goal was to improve the accuracy, reliability, and scalability of the client’s ASO off-target prediction model. They wanted to reduce false positives, increase confidence in candidate prioritization, and minimize the experimental burden. To achieve this, the client required advanced bioinformatics solutions—leveraging machine learning, mismatch pattern analysis, and RNA-seq–based validation—to refine prediction algorithms and establish a robust framework for RNA therapeutic development.
Our approach
With no predefined framework in place, we began by working with the client to define a clear roadmap, laying out each stage of analysis and delivery. This roadmap was developed in close collaboration and iteratively refined with the client’s scientific informatics team.
Excelra’s strategy focused on three pillars:
Exploratory Pattern Mining
We conducted in-depth data mining of ASO–transcript alignments to uncover statistical and positional patterns that influence off-target effects. We applied statistical methods such as log-odds ratios (a measure of mismatch enrichment) along with custom heatmaps to visualize positional effects. These analyses identified mismatch positions most predictive of transcript downregulation, directly informing the refinement of off-target prediction rules and model features.
Pipeline Enhancement
To enhance off-target detection, we benchmarked multiple alignment tools and parameter configurations. By selecting a more adaptable aligner and implementing a custom patch script, we eliminated the mismatch threshold constraint of the existing pipeline—enabling broader transcriptome coverage without disrupting downstream systems.
Interpretable ML Modeling
We developed machine learning models powered by features extracted from alignment data—such as mismatch positions and sequence alterations—to predict off-target activity with greater precision. Using SHAP (SHapley Additive exPlanations) analysis and feature importance methods, we ensured model interpretability, enabling clear insights into mismatch tolerance patterns. These findings not only enhanced predictive performance but also provided data-backed validation and refinement of the client’s existing detection rules.
We delivered all analytical outputs through Quarto notebooks, ensuring full transparency, reproducibility, and ease of knowledge transfer. The workflow was seamlessly integrated into the client’s high-performance computing environment and Snakemake-based pipelines, enabling immediate usability within their existing infrastructure and supporting long-term scalability through scientific data management.
Our solution
Uncovering Mismatch Patterns through Iterative Statistical Analysis
In response to evolving client questions and the availability of new RNA-seq analysis data, we adopted an agile, iterative approach—revisiting and expanding our pattern analysis to refine hypotheses and validate findings.
Through successive rounds of statistical exploration and pattern analysis, we deepened our understanding of mismatch-position effects and ensured the consistency and robustness of observed trends.
We designed and executed four sets of analysis and twelve visualization types, applied to two large RNA-seq analysis datasets covering 134 ASOs.
One type of analysis successfully revealed patterns in mismatch frequency and position linked to potential off-target effects. We compared mismatch patterns in downregulated transcripts to their overall background frequency, using enrichment-based methods to identify positions and configurations more likely to be associated with off-target effects. The results were visualized as heatmaps, allowing intuitive interpretation of positional effects across large ASO datasets.
Additionally, we profiled ASO binding preferences across transcript regions—assessing positional distribution across exons and introns—to uncover regional susceptibility to off-target effects.
Together, these provided a strategic foundation for enhancing the client’s predictive model, informing rational ASO design, and guiding experimental prioritization with greater confidence.
Results and Impact
Excelra’s advanced bioinformatics solutions and ML-driven approach delivered measurable improvements across all key performance areas:
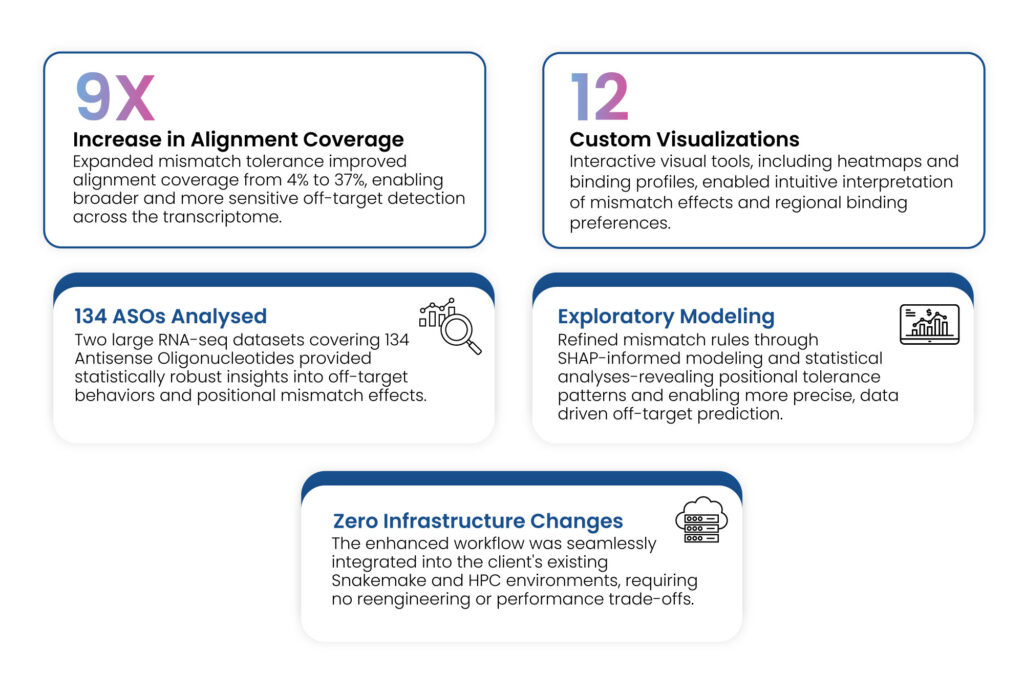
- 9x Increase in Alignment Coverage
- 12 Custom Visualizations
- 134 ASOs Analyzed through RNA-seq analysis
- Exploratory Modeling with SHAP
- Zero Infrastructure Changes
These outcomes accelerated the client’s ASO screening process, improved predictive precision, and empowered experimental teams with interpretable, data-backed insights, laying a scalable foundation for RNA therapeutics development.
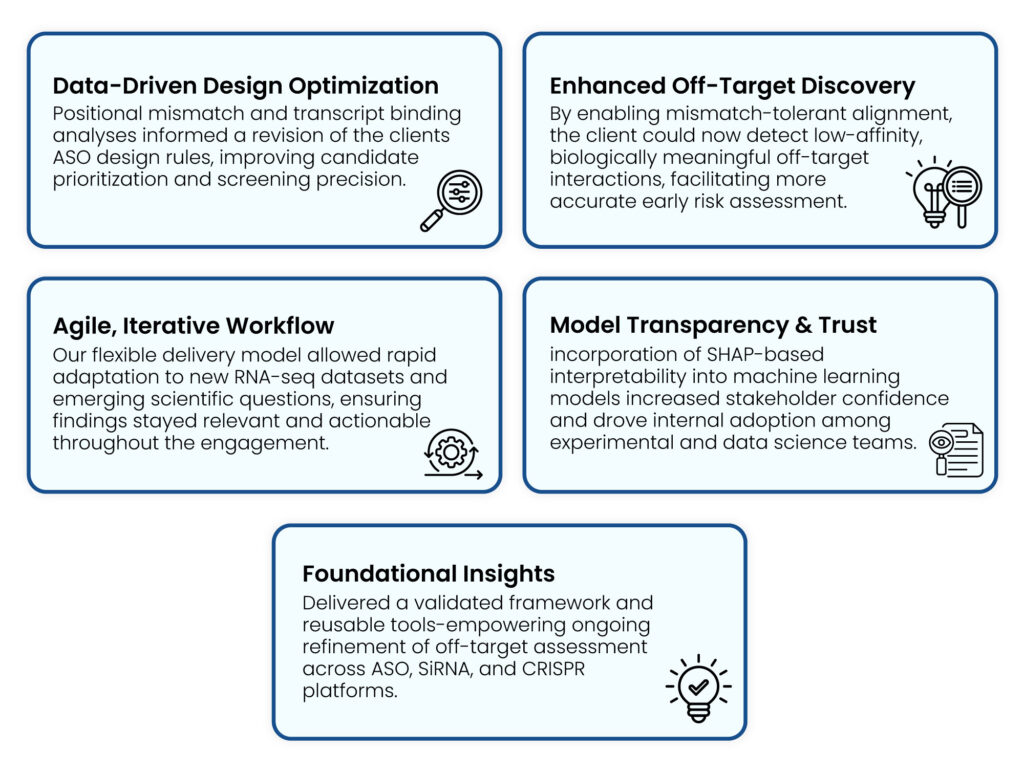
Strategic Benefits:Excelra’s integrated approach not only addressed technical bottlenecks but also delivered long-term strategic value for the client’s RNA therapeutics pipeline:
- Data-Driven Refinement of ASO Design Rules
Statistical analysis and interpretable ML uncovered mismatch patterns that informed refinements to existing detection guidelines—laying the groundwork for more precise off-target prediction frameworks.
- Expanded Alignment and Off-Target Discovery
Overcoming mismatch threshold constraints led to a 9x increase in alignment coverage (from 4% to 37%), enabling broader transcriptome screening and identification of low-affinity interactions.
- Improved Interpretability and Scientific Trust
SHAP-based analysis provided transparency into model outputs, helping scientists better understand mismatch tolerance and supporting informed experimental design.
- Agile, Iterative Exploration Aligned with Evolving Needs
The team adapted quickly to new RNA-seq data and evolving questions—revisiting hypotheses and refining insights through collaborative cycles.
- Foundational Insights for Scalable RNA Therapeutics
While formal rule changes were deferred pending further validation, the project provided a validated framework and reusable tools for ongoing refinement across ASO, siRNA, and CRISPR modalities.
By integrating FAIR data principles, robust scientific data management, and cheminformatics pipelines, this project established a scalable framework applicable across siRNA and CRISPR modalities.
Why Excelra?
Excelra combines deep scientific expertise with cutting-edge data science to empower confident, data-driven decisions across drug discovery and development. Our unique blend of capabilities ensures impactful outcomes tailored to each client’s scientific challenge.
With Excelra, clients don’t just get a service provider; they gain a scientific partner committed to measurable impact and long-term success.
Conclusion
With no established solution in place, we delivered both the strategic roadmap and hands-on execution to address a complex off-target prediction challenge in RNA therapeutics. Excelra’s work led to a 9x increase in alignment coverage, analysis of 134 ASOs across two large RNA-seq analysis datasets, and the creation of a scalable, mismatch-tolerant pipeline. By combining bioinformatics solutions, scientific informatics, and advanced scientific data management aligned with FAIR data principles, the client gained a reproducible framework that accelerated ASO design and strengthened future therapeutic pipelines with cheminformatics integration.

