Overview
This case study highlights our robust RNA-seq analysis pipeline for patient-derived xenograft (PDX) models to derive biological insights in cancer biology. Using advanced QC, demousing, and bioinformatics, we enabled the identification of actionable biomarkers for therapy and survival in multiple cancer models.

Our client
A US-based precision medicine biotech company focused on oncology research partnered with us to extract deeper insights from their PDX (patient-derived xenograft) RNA-seq data. They sought a trusted data science and bioinformatics partner capable of handling large-scale sequencing datasets, identifying therapeutic biomarkers, and integrating survival analysis for translational research applications.

Client’s challenge
The client had raw RNA-seq data from PDX ( patient-derived xenograft ) models containing mouse stromal RNA contamination.
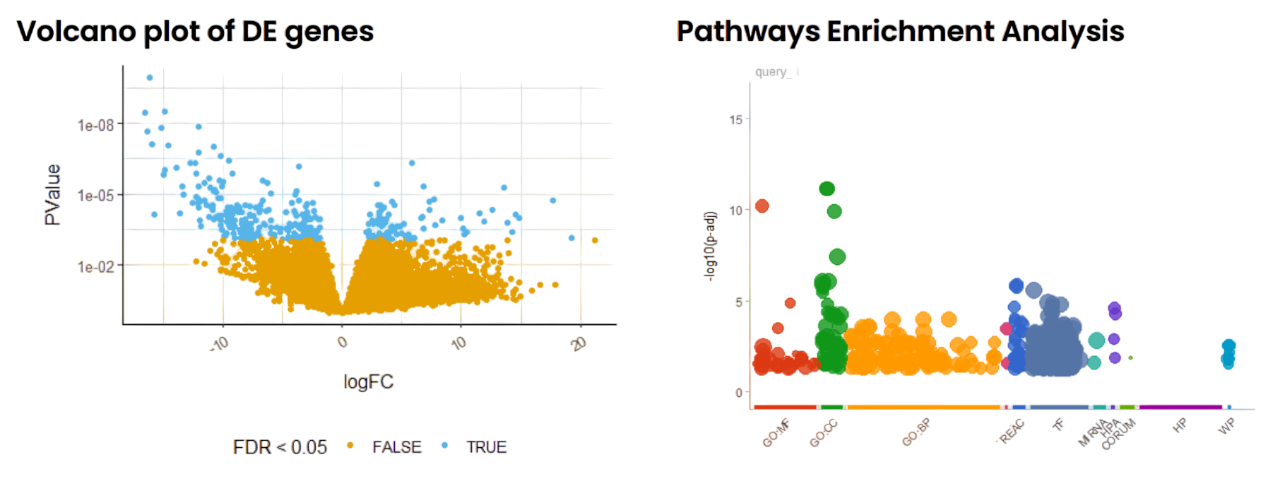
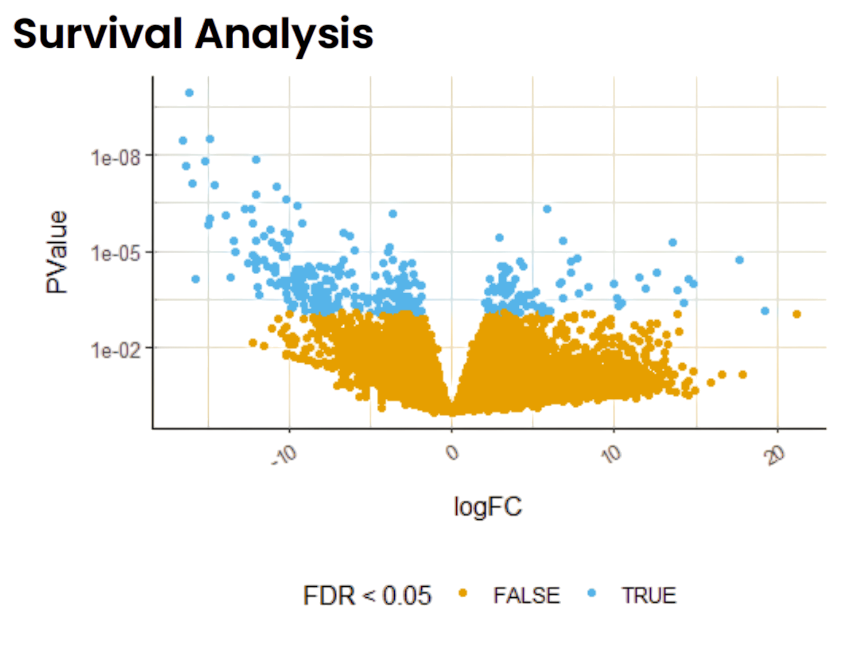
They needed to isolate human tumor-specific reads, analyze gene expression changes, and understand disease mechanisms via Volcano plots, survival analysis, and pathway enrichment.
The goal was to support biomarker discovery within a cancer biology framework.

Client’s goals
- Demouse RNA-seq FASTQ files from patient-derived xenograft models.
- Ensure data integrity using robust QC tools.
- Identify differentially expressed genes via an automated RNA-seq analysis pipeline.
- Visualize and interpret DEGs through Volcano plots and pathway enrichment.
- Integrate survival data for prognostic biomarker identification in various cancer models.
Our approach
We developed an end-to-end RNA-seq pipeline, combining automated demousing, QC, quantification, and statistical modeling:
Data Quality Control-
FASTQC, MultiQC, RSeQC, Picard, Preseq, dupRadar, and Qualimap ensured reliable RNA-seq inputs.
Demousing with Xengsort-
Removed mouse RNA, isolating only human tumor reads from PDX models.
Alignment & Quantification-
STAR aligner and RSEM generated normalized count tables across cancer types.
Differential Expression Analysis-
Used DESeq2, edgeR, and limma to find DEGs driving tumor behavior.
Volcano Plots & Enrichment-
Visualized DEGs and enriched pathways (ORA, GSEA) in cancer biology.
Survival Integration-
Linked expression data with patient outcome for biomarker discovery.

Our solution
- Delivered demoused count matrices for each cancer model.
- Generated clear Volcano plots showing statistically significant DEGs.
- Mapped genes to critical pathways related to tumor progression and treatment resistance.
- Uncovered potential prognostic biomarkers via integrated survival analysis.