Overview
This case study demonstrates Excelra’s comprehensive RNA-seq data analysis pipeline applied to public cancer datasets for small cell lung carcinoma (SCLC) and ovary carcinoma (OC). Using advanced data curation, transcriptomic analysis, and gene annotation, we identified and prioritized lineage-specific targets through integrated bulk and scRNA data.

Our client
A research-driven oncology-focused organization aimed to identify tumor-enriched and tissue-specific therapeutic targets for precision oncology initiatives in SCLC and OC. They needed a robust pipeline capable of analyzing large-scale RNA data from public repositories and applying advanced bioinformatics for target prioritization.

Client’s challenge
- Identify reliable tumor-specific and lineage-specific targets from heterogeneous transcriptomic datasets.
- Integrate and normalize RNA-seq data from multiple sources (TCGA, GEO, scRNA).
- Remove batch effects and perform RNA-seq data analysis at both bulk and single-cell levels.
- Annotate genes for druggability and relevance to cancer biology using comprehensive gene annotation methods.

Client’s goals
- Perform target identification using bulk RNA-seq and scRNA data for SCLC and OC.
- Apply lineage-based filtering using vital organ datasets to remove off-target genes.
- Prioritize targets using pathway analysis and gene annotation databases.
- Generate actionable insights for downstream precision oncology validation
Our approach
We curated and integrated public cancer datasets from TCGA and GEO, focusing on RNA-seq data from tumor and matched normal samples. We performed normalization and batch correction, followed by comparative expression analysis to identify tumor-enriched genes. We used scRNA-seq data to refine targets based on cell-type-specific expression.
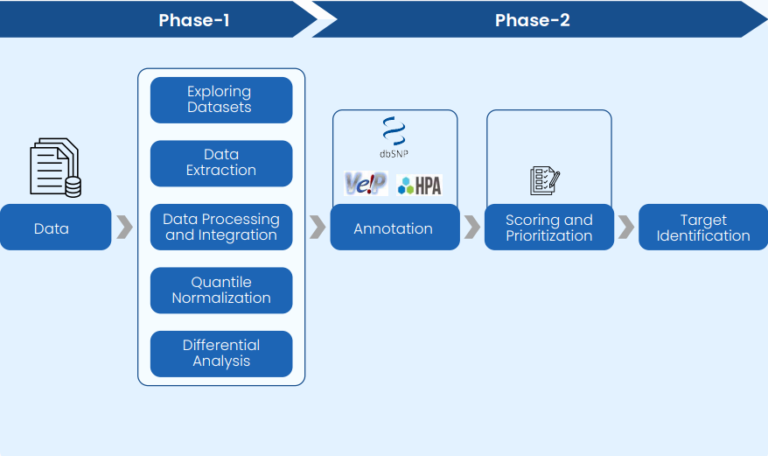
Data Curation & Integration-
Publicly available cancer datasets from TCGA and GEO were collected for small cell lung carcinoma (SCLC), ovary carcinoma (OC), and their respective normal tissue controls. scRNA-seq data for tumor and normal tissues (lung, ovary, and other vital organs) were also integrated to strengthen the transcriptomic analysis. This phase also included structured data curation to ensure compatibility and accuracy.
Normalization & Batch Correction-
Quantile normalization and batch effect removal were applied across datasets to ensure uniformity and reduce technical variance in RNA data from diverse sources.
Target Identification-
- Tumor Enrichment Analysis revealed differentially upregulated genes specific to SCLC and OC compared to their respective normal tissues using our RNA-seq data analysis pipeline.
- Lineage-Specific Target Analysis was performed by comparing cancer datasets with vital normal organ data to filter out broadly expressed genes.
- scRNA-based Analysis allowed precise identification of tumor-restricted genes for SCLC and OC by comparing expression at single-cell resolution against lineage-defining normal cells.
Annotation & Prioritization-
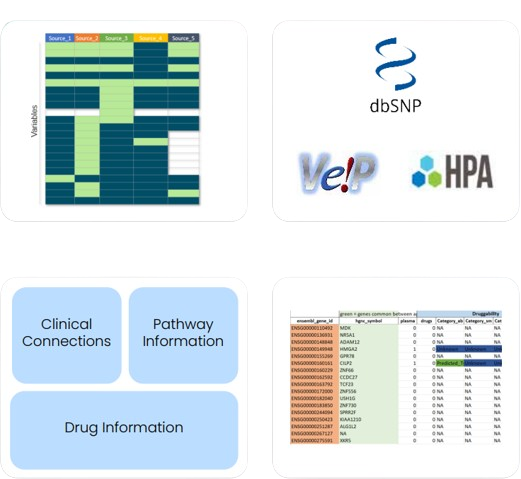
Identified genes were annotated using multiple biological and pharmacological databases. Gene annotation was supported by pathway involvement and known drug interactions. Target prioritization was based on expression specificity, pathway relevance, druggability, and overall clinical potential within precision oncology.
Deliverables Generated-
- A curated list of tumor-enriched and lineage-restricted targets for SCLC and OC backed by detailed transcriptomic analysis.
- Prioritized gene sets for further preclinical validation.
- Detailed gene annotation of function, expression profiles, known drugs (if any), and disease relevance.
- Final report and structured data files for downstream use and integration into scientific applications.
Conclusion
By leveraging a structured pipeline integrating bulk and single-cell transcriptomic data, Excelra successfully identified and prioritized tumor-enriched and lineage-specific targets for small cell lung carcinoma and ovary carcinoma. The dual-layered approach—combining differential expression and lineage filtration—ensured the selection of high-confidence targets with minimal off-target risk in vital organs. The final output provides a robust foundation for downstream experimental validation and therapeutic development in precision oncology.

