Overview
A leading research organization in the pharmaceutical and agricultural sector wanted to map the epigenetic landscape of the complex hexaploid wheat genome. Their goal was to uncover gene regulation mechanisms influencing traits like drought tolerance, disease resistance, and yield. They required an advanced computational framework to integrate multi-omics data, analyze epigenetic modifications, and map DNA loops.

Our client
The client is a Germany-based organization engaged in agricultural and pharmaceutical research. Their focus was on developing a deeper understanding of epigenetic mechanisms to drive innovation in crop improvement and functional genomics.

Client’s challenge
To enable deep insights into wheat gene regulation, the client needed an advanced computational biology solution capable of managing multi-omics data analysis. The complexity of the hexaploid wheat genome posed significant barriers to analyzing chromatin structure, histone modifications, and transcriptional regulation. Moreover, organizing and aligning data from disparate sources called for rigorous data curation and FAIR data practices.

Client’s goals
Their aim was to develop a scalable computational analysis pipeline that integrates high-throughput sequencing data for identifying genome-wide histone modifications and chromatin loops. Additionally, they sought to leverage FAIR data principles for future scalability and reproducibility of research insights.
Our approach
Integration of multi-omics datasets
- Leveraged high-throughput sequencing data, including Hi-C and ChIP-Seq, to capture chromatin interactions and histone modifications.
- Combined multiple epigenetic data types for comprehensive multi-omics data analysis of wheat genome regulation.
Computational framework development
- Designed a robust computational biology pipeline for Hi-C and ChIP-Seq data processing, optimized for polyploid genomes.
- Applied algorithms for genome-wide computational analysis of histone modifications and DNA loops.
- Implemented FAIR data standards and scalable workflow design for reproducibility.
De Novo analysis of hexaploid wheat genome
- Developed a novel approach to analyze Hi-C data in wheat, overcoming polyploid-specific challenges.
- Achieved high-resolution chromatin interaction maps to support accurate interpretation of gene regulatory elements through integrated multi-omics data analysis.
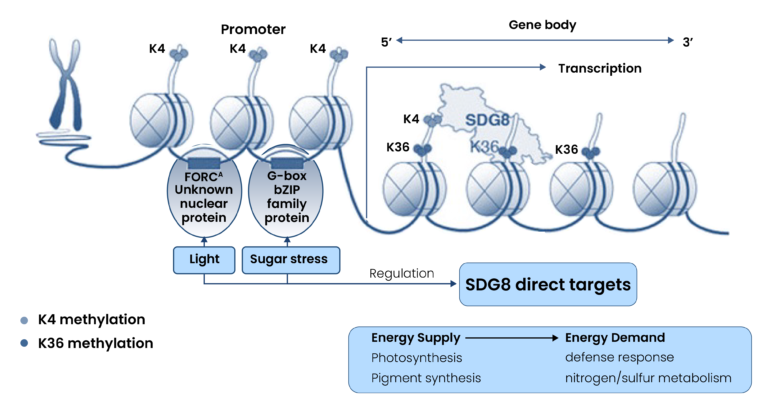
Our solution
By integrating cutting-edge computational analysis tools and structured data curation workflows, we built a scalable framework that allowed rapid and precise analysis of histone modifications and chromatin loops. The Hi-C pipeline, optimized for polyploid wheat genomes, demonstrated the potential of combining computational biology and FAIR data practices in epigenetics research.

Conclusion
The client benefited from a reduction in analysis time of over 80%, cutting the process from 21 weeks to just 4 weeks. This transformative result accelerated their research timelines and established them as a pioneer in agricultural epigenetics. With a scalable computational framework and robust multi-omics data analysis capabilities, they are now positioned to advance the understanding of gene regulation through histone modifications and epigenetic patterning in crop genomes.

