Overview
Excelra partnered with a leading Computational Oncology department to develop customized, production-ready bioinformatics pipelines tailored for RNA-Seq, scRNA-Seq, WGS/WES, and HLA typing data. The objective was to handle complex biological datasets and enhance research throughput. Addressing challenges in data complexity, performance optimization, and tool integration, Excelra implemented a modular, scalable architecture using Nextflow and Docker. The phased rollout from Q2 2023 to Q1 2024 included continuous enhancements in error handling, QC, and internalization of Sarek workflows. This initiative empowered oncology researchers with high-performance, reproducible pipelines, significantly accelerating biomarker discovery and genomic analyses.

Our client

Client’s challenge
- Data complexity: Managing diverse biological data formats and structures.
- Algorithm selection: Choosing suitable tools and algorithms for analysis tasks.
- Performance optimization: Ensuring efficiency in speed, resource usage, and scalability.
- Integration challenges: Integrating multiple tools and modules while maintaining
compatibility.

Client’s goals
Develop and implement robust pipelines tailored to each data-type for comprehensive analysis. Additionally, provide essential bioinformatics support to enhance the capabilities of the Computational Oncology department in conducting cutting-edge research.
Our approach
Data complexity
Managing diverse biological data formats and structures.
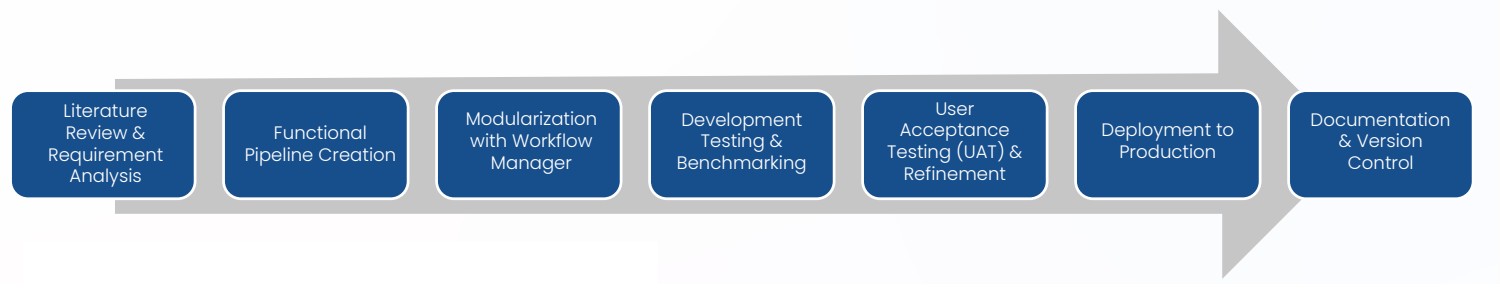
Input: Data types
- RNA-Seq
- scRNA-Seq
- WGS
- WES
Deliverables: Production ready pipelines
- RNA-Seq pipeline (Single and paired-end)
- scRNA-Seq pipeline (single end and paired end)
- HLA typing pipeline
- Germline WGS/WES pipeline
- SAREK pipeline (Germline, somatic and Tumor)
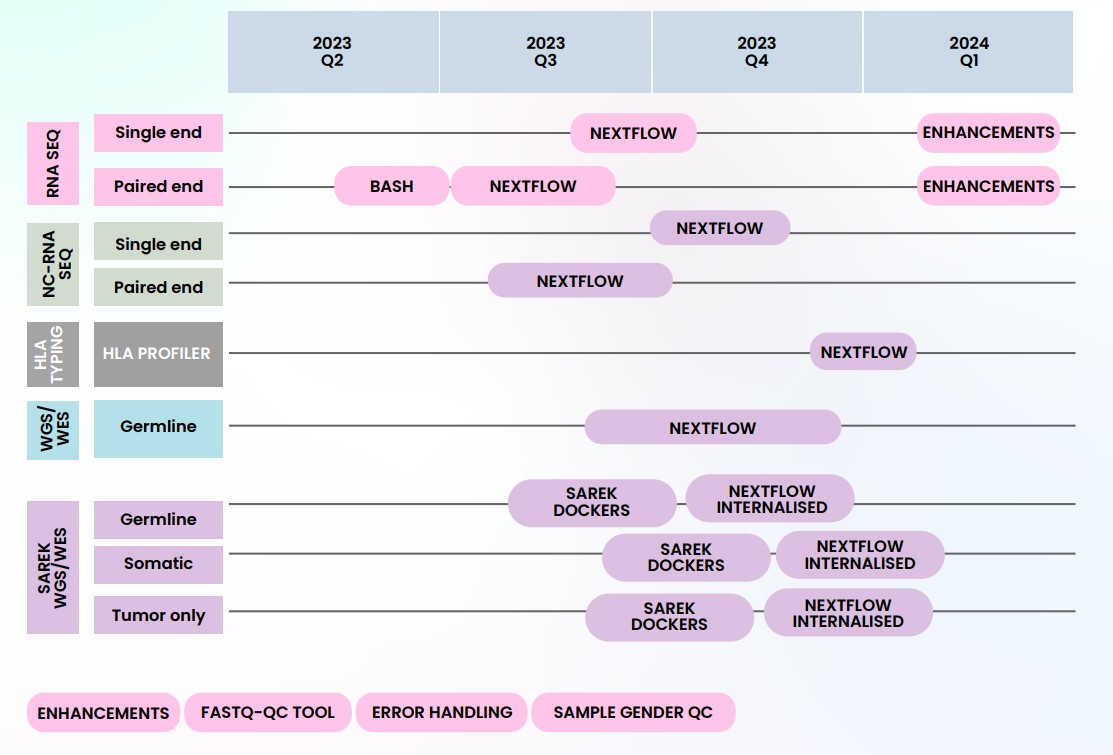
Figure: Implementation timeline

Impact and results
- RNA-Seq (single-end and paired-end)
- scRNA-Seq (single-end and paired-end)
- Whole Genome Sequencing (WGS)
- Whole Exome Sequencing (WES)
- HLA typing
- SAREK pipeline for germline, somatic, and tumor variant calling

