Overview
Excelra partnered with a leading global health and nutrition company to accelerate microbiome research and strain engineering using customized bioinformatics pipelines. The project focused on optimizing biomolecule production, enhancing strain development, and automating bioinformatics workflows for reproducible, high-throughput analysis. By leveraging advanced synthetic biology approaches and end-to-end bioinformatics pipelines, the client was able to achieve faster insights and greater operational efficiency.

Our client
An innovation-first health and nutrition company based in Belgium with a strong emphasis on microbiome research. Their operations include strain development, probiotic design, and precision nutrition, where bioinformatics pipelines and data-driven workflows play a central role in R&D.

Client’s challenge
The client aimed to enhance biomolecule production through precision strain engineering while improving microbiome research efficiency. Their key challenges included:
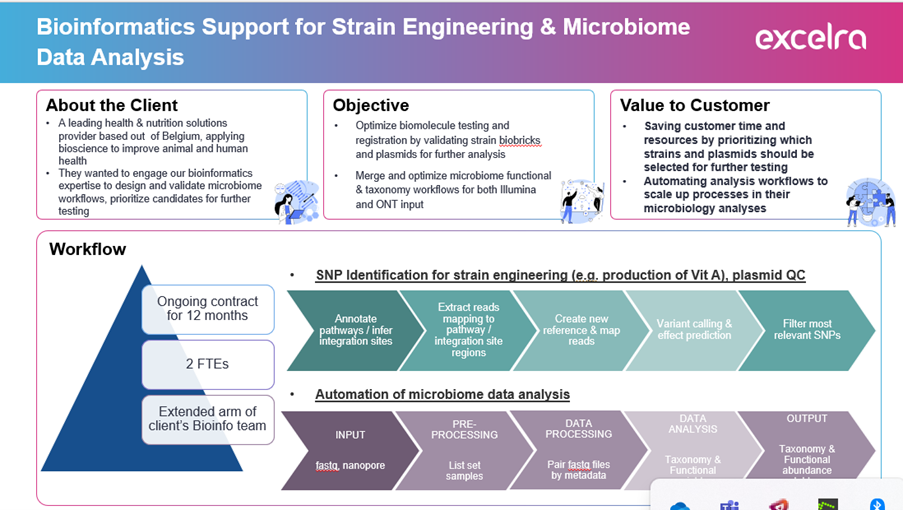
- Identifying genetic variations for metabolic optimization, such as Vitamin A biosynthesis.
- Validating engineered plasmid constructs and custom strain biobricks.
- Developing a scalable, automated bioinformatics workflow compatible with both Illumina (short-read) and Oxford Nanopore Technologies (long-read) sequencing platforms.
- Reducing analysis time without compromising accuracy or flexibility in strain engineering workflows.

Client’s goals
The client sought to:
- Build automated bioinformatics pipelines for reproducible microbiome research and strain engineering.
- Increase throughput and efficiency in strain development and experimental validation.
- Enable precision synthetic biology applications through reliable genetic construct validation.
Our approach
Excelra designed a tailored, end-to-end bioinformatics pipeline to meet the client’s scientific and operational goals, focusing on:
- SNP Discovery for Strain Engineering: Developed bioinformatics pipelines to detect key genetic variants influencing biosynthetic pathways, particularly for Vitamin A.
- Plasmid Construct & Biobrick Validation: Implemented workflows to verify synthetic biology constructs and engineered strains.
- Taxonomic and Functional Analysis: Created robust bioinformatics workflows capable of handling both Illumina and ONT sequencing data to support microbiome research.
- Workflow Automation: Automated repetitive analysis tasks to enable high-throughput, reproducible bioinformatics pipelines.
For more details on Excelra’s offerings, see our computational biology services.
Our solution
Excelra delivered a scalable, automated solution that enabled the client to:
- Accelerate biomolecule discovery and strain optimization through targeted genetic analysis.
- Implement automated bioinformatics pipelines to reduce turnaround times.
- Prioritize high-potential strains and constructs for rapid experimental validation.
- Enhance reproducibility and scalability across microbiome research projects.
Conclusion
Through this collaboration, the client transformed their microbiome research and strain engineering programs. Key achievements included:
- 40% reduction in analysis turnaround time, enabling faster decision-making.
- 60% improvement in data analysis efficiency via automated bioinformatics pipelines.
- 25% increase in high-potential genetic variant identification through targeted SNP discovery.
The automated bioinformatics workflows, combined with synthetic biology strategies, positioned the client for next-generation microbiome research and precision strain engineering. Excelra’s expertise ensured faster insights, operational flexibility, and a foundation for future-ready synthetic biology innovation.

