Overview
Excelra partnered with a leading agri-food company to deliver biomarker discovery solutions for accurate chicken cell line differentiation. By leveraging computational biology services and advanced data curation, we enabled faster and cost-effective lineage validation compared to whole genome sequencing.Management.

Our client
An innovation-driven European agri-food manufacturer engaged in genomic analyses of animal-derived cell lines for research, development, and production validation. They place strong emphasis on traceability, reproducibility, and biological lineage fidelity across their processes.

Client’s challenge
A leading food and beverage manufacturing company required a robust, scalable solution to differentiate between multiple chicken (Gallus gallus) cell lines. Each cell line originated from a distinct embryo, and accurate lineage tracking was essential for quality control, regulatory compliance, and scientific reproducibility.
Their existing method—Whole Genome Sequencing (WGS)—was highly accurate but time-intensive, costly, and impractical for routine application. Additionally, their workflow lacked integration of computational biology methods and systematic data curation, limiting deeper biological insights.

Client’s goals
The client’s goal was to get a faster, cost-effective approach that maintained the precision of WGS while being suitable for frequent use. They sought to leverage modern computational biology approaches and reliable data curation to build a more scalable quality control framework.
Our approach
Excelra employed a multi-stage bioinformatics pipeline to deliver lineage-specific genetic biomarkers. Our methodology included:
- Whole Genome Sequencing (WGS) Analysis of multiple chicken cell lines to capture a comprehensive variant landscape.
- Advanced Variant Calling and Filtering to identify high-confidence SNPs, SNVs, and indels, ensuring only robust markers were retained.
- Customized Bioinformatics Workflow integrating population genetics and lineage inference to correlate variants with biological ancestry.
- Biomarker Prioritization supported by systematic data curation to identify targets suitable for PCR-based assays, drastically reducing the need for repetitive WGS.
Similar approaches have been highlighted in our Identification of Genomic Biomarkers for Cell Line Differentiation case study.
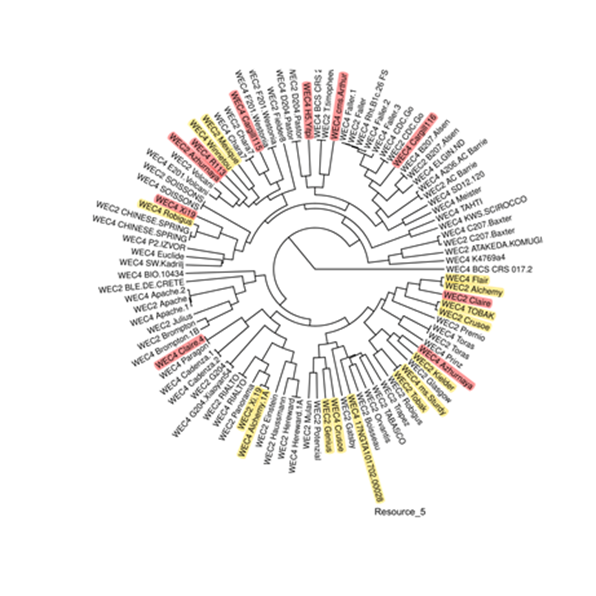
Our solution
Excelra delivered a cost-effective, high-throughput solution to enable routine quality control without compromising accuracy:
- PCR-Validatable Biomarkers Identified- Empowered rapid screening of cell lines without full genome sequencing.
- Reduced Operational Costs- Transitioned QC from genome-wide to targeted variant analysis.
- Improved Traceability and Lineage Fidelity across production batches.
This integrated solution leveraged advanced computational biology and in-depth data curation, enabling a scalable and regulatory-ready framework powered by Excelra’s domain expertise and GOSTAR™ Database.
Conclusion
Excelra’s biomarker discovery approach enabled a >90% reduction in turnaround time compared to whole genome sequencing, while maintaining >98% accuracy in cell line differentiation. By identifying a panel of PCR-validated genetic markers, we eliminated the need for repetitive WGS in routine quality control—cutting operational costs by over 70%.
The streamlined pipeline enhanced lineage traceability across batches, improved regulatory readiness, and empowered the client to scale QC throughput by 5x, significantly accelerating decision-making in R&D and production environments. This success highlights the impact of computational biology and high-quality data curation in advancing biomarker discovery.

