Overview
This case study showcases how Excelra helped a leading clinical diagnostics lab specializing in genetic testing streamline their in-house Next-Generation Sequencing (NGS) workflows. By transitioning from traditional batch-based architecture to a modern, modular NGS pipeline using Nextflow, the client significantly improved sample-level processing, reduced turnaround time, and enhanced overall pipeline scalability. The solution aligned with best practices in genome analysis, clinical diagnostics, and precision medicine. The engagement exemplifies Excelra’s capabilities in scientific workflow optimization, with supporting services such as Computational Biology and Scientific Data Management.

Our client
A US-based leading clinical diagnostics laboratory focused on genetic testing, leveraging high-throughput Next-Generation Sequencing (NGS) technologies to deliver precision medicine solutions. The client had their existing NGS pipelines for data processing and analysis done in a batch-processing manner, which increased the turnaround time (TAT) and led to inefficiencies due to a high volume of incoming samples. The client wanted us to review and optimize their existing architecture to implement sample-based workflows that would reduce TAT and improve performance.

Client’s challenge
A US-based clinical diagnostics laboratory specializing in genetic testing was facing significant challenges in optimizing the performance of its in-house NGS pipelines. The client received multiple patient samples individually and had a series of benchmarked pipelines for job management and analytical validations. Most were developed in Python, while some were in Nextflow, operating in a batch-processing manner. This structure caused delays, as individual samples had to wait for an entire batch to accumulate before analysis could begin—impacting high-priority cases in particular. The team wanted to implement sample-level processing, along with batch correction to ensure consistent and scalable genome analysis workflows.

Client’s goals
The client’s goals were centered on improving efficiency, reducing processing delays, and enabling better scalability through automation. Specific objectives included:
- Implementing sample-based NGS workflows
- Modularizing and refactoring pipeline scripts
- Improving maintainability and processing speed
- Ensuring high-quality documentation
- Supporting precision medicine diagnostics with faster data availability
Our approach
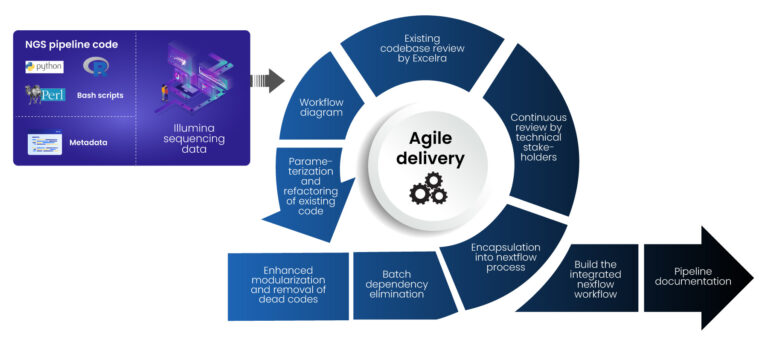
Excelra adopted an agile approach to address the client’s challenges systematically. First step was to identify bottlenecks and inefficiencies in the existing NGS pipeline. A comprehensive assessment was initiated for their Carrier Screening, Hereditary Cancer, WES and WGS pipelines, all of which play critical roles in modern clinical diagnostics and precision medicine.
- Reviewing the existing codebase to understand workflow dependencies and inefficiencies and substitute fixed values with variables, and transfer them to a configuration file, remove any redundant or inactive code in the codebase and creating the code workflow that provides associations and help build causalities between the scripts. This process is essential to optimize high-throughput Next-Generation Sequencing (NGS) systems used in genome analysis workflows.
- Developing a workflow diagram of codes/scripts to visualize dependencies and streamline execution—an important component in scalable NGS pipelines.
- Refactoring and parameterizing scripts to improve modularization, remove redundant code, and enhance maintainability—crucial for sustainable precision medicine platforms.
- Eliminating batch dependencies within each method to enable a shift from batch-based processing to a sample-specific approach, ensuring reduced turnaround time and faster clinical diagnostics outcomes.
- Encapsulating processes into Nextflow modules for enhanced workflow automation and scalability, enabling the client to future-proof their NGS workflows.
- Conducting continuous review cycles with the client’s technical team to ensure alignment with their objectives, while maintaining high standards of reproducibility in genome analysis.
- Building an integrated Nextflow workflow by knitting modules in workflows and sub-workflows to unify all optimized processes. This integration supports end-to-end performance and is aligned with Excelra’s expertise in Scientific Application Development for Drug Discovery.
- Creating high-quality documentation to ensure reproducibility and future scalability, in line with FAIR Data Principles that support compliance, traceability, and collaborative genome analysis across projects.
Our solution
Excelra successfully implemented an optimized NGS pipeline using Nextflow, bringing several operational benefits for the client:
Sample-based processing
Upgraded the Next-Generation Sequencing (NGS) pipelines for a sample-based processing as opposed to previous batch processing. This helped to generate all clinical diagnostics reports on a per-sample basis, supporting greater precision in genome analysis.
Increased efficiency and scalability
The modularized pipeline improved processing speed by 20% and eliminated workflow bottlenecks, enabling smoother execution of genetic testing procedures used in precision medicine workflows.
Code maintainability and refactoring
Parameterized scripts made it easier for the client to update and manage their workflows. This aligns with best practices in Scientific Application Development and long-term sustainability of NGS pipelines.
Reduced turnaround time (TAT)
Transitioning from batch processing to sample-specific analysis significantly cut down processing delays and reduced the TAT to approximately 3 hrs. This supports faster, more efficient delivery of results in clinical genomics settings.
High-quality documentation standards
Comprehensive documentation ensured that future modifications could be made seamlessly. It also ensured compliance with FAIR data principles, enhancing data transparency and reusability in bioinformatics environments.
Deliverables
- A fully optimized Nextflow workflow for each individualized pipeline.
- Refactored and parameterized scripts to improve modularization.
- Validation of results to ensure accuracy and efficiency in the new workflow.

Conclusion
By implementing Nextflow and modular architecture, Excelra enabled the client to transform their batch-based NGS pipelines into efficient, scalable workflows that align with clinical diagnostics needs and precision medicine standards. The optimized infrastructure supported higher sample volumes, accelerated reporting timelines, and ensured adaptability for future pipeline enhancements. The engagement reinforces Excelra’s ability to deliver end-to-end scientific informatics solutions for evolving challenges in genome analysis and Next-Generation Sequencing.

