Overview
This case study focuses on enhancing the client’s genomic reference data repository using advanced bioinformatics tools, ensuring clinical genomics workflows benefit from traceable, immutable identifiers. By incorporating FAIR data principles and robust data versioning strategies, Excelra significantly improved the integrity and accessibility of the client’s reference data for advanced DNA sequence analysis and precision oncology initiatives.

Our client
A leading companion diagnostic company based in the US, specializing in oncology and precision medicine, and known for delivering bioinformatics tools for clinical research and diagnostic solutions. Their existing systems lacked the capability to effectively trace genomic reference versions, affecting the accuracy of their DNA sequence analysis pipelines and limiting clinical genomics insights.
This limitation also impacted the implementation of FAIR data principles, critical for ensuring data transparency, interoperability, and reusability in a highly regulated precision oncology space.
For more information on our genomic and computational offerings, explore our Computational Biology Services and Scientific Data Management solutions.

Client’s challenge
The client’s bioinformatics tools pipelines required a wide array of reference file types such as BED, GTF, and FASTA formats. However, due to the absence of data provenance and traceability, scaling their clinical genomics and molecular analytics efforts became a major challenge. They could not reliably map specific molecular findings back to a fixed version of the genomic reference data, leading to concerns around data consistency in precision oncology diagnostics.
Additionally, the current tooling lacked the support for managing genomic interval data files with immutable identifiers, a core requirement in upholding FAIR data principles.

Client’s goals
- To enhance their open-source tooling and integrate a robust genomic reference storage system.
- To enable unique UUID-based tracking of interval data files (BED, GTF, etc.) for streamlined DNA sequence analysis.
- To ensure adherence to FAIR data principles, making data traceable, reproducible, and accessible across the organization.
- To scale and secure their clinical genomics pipelines by adopting better data governance practices.
These goals aligned with the core objectives of precision oncology, where accurate reference data forms the backbone of patient-specific diagnostics and therapeutic strategies
Our approach
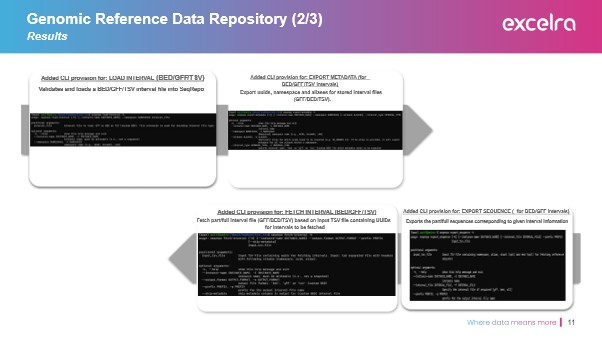
Objective 1: Data Fetching & Extraction
The team implemented a system to process various genomic file types (FASTA, GTF/GFF, BED, TSV) by indexing and storing them using SQLite, thereby standardizing the input for bioinformatics tools and enabling downstream DNA sequence analysis workflows.
Objective 2: Data Storage & Processing
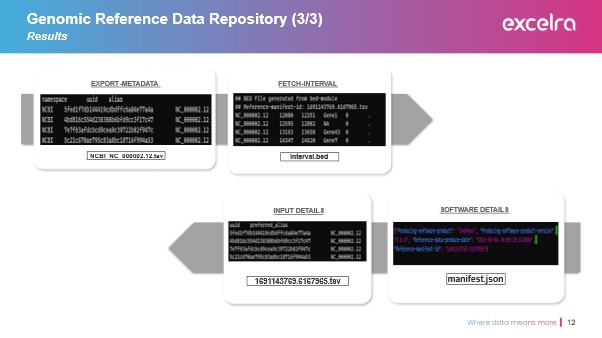
We assigned each genomic interval a unique UUID, ensuring strict traceability in line with FAIR data principles. Files were renamed and indexed, allowing the generation of complete reference genome sets from collated identifiers. The structured outputs included:
- A consolidated genomic file in the required format
- A UUID manifest file
- A JSON metadata file capturing tool version, reference manifest ID, and timestamps
This streamlined approach supported advanced analytics within clinical genomics and provided a strong foundation for scaling precision oncology operations.
Our approach is comparable to strategies seen in our Ideate, Design and Develop a Gene Visualization Platform case study, where seamless genomic data management played a pivotal role.
Technologies Used:
- Python-based SeqRepo enhancements
- Docker deployment for portability
- Enhanced command-line utilities for genomic reference data management
- Integration of FASTA, BED, GTF/GFF, and TSV file handling
Our solution
- Integrated open-source platforms to enable secure genomic data storage with immutable, UUID-based identifiers—essential for clinical genomics workflows.
- Extended functionalities for bulk downloading and generation of complete human genome reference sets—accelerating DNA sequence analysis.
- Introduced management capabilities for genomic interval data (BED, GFF) tied to consistent identifiers—ensuring alignment with FAIR data principles.
To ensure scalability, portability, and future enhancements, the solution utilized Docker-based deployment and enhanced CLI utilities. This solution aligns with Excelra’s broader offerings in Scientific Application Development and data-intensive bioinformatics tools ecosystems.

Conclusion
Excelra’s enhancements to the SeqRepo platform ensured accurate genomic reference versioning and immutable data handling—delivering a scalable, auditable, and efficient solution for precision oncology diagnostics. The upgraded system empowered the client’s research and diagnostics teams to conduct more reliable DNA sequence analysis, enforce FAIR data principles, and elevate their clinical genomics capabilities across various disease areas.

